Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast
- PMID: 10880460
- PMCID: PMC313942
- DOI: 10.1093/emboj/19.13.3475
Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast
Abstract
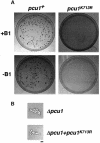
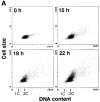
A ubiquitin-like modifier, NEDD8, is covalently attached to cullin-family proteins, but its physiological role is poorly understood. Here we report that the NEDD8-modifying pathway is essential for cell viability and function of Pcu1 (cullin-1 orthologue) in fission yeast. Pcu1 assembled on SCF ubiquitin-ligase was completely modified by NEDD8. Pcu1(K713R) defective for NEDD8 conjugation lost the ability to complement lethality due to pcu1 deletion. Forced expression of Pcu1(K713R) or depletion of NEDD8 in cells resulted in impaired cell proliferation and marked stabilization of the cyclin-dependent kinase inhibitor Rum1, which is a substrate of the SCF complex. Based on these findings, we propose that covalent modification of cullin-1 by the NEDD8 system plays an essential role in the function of SCF in fission yeast.
Figures










Similar articles
-
Two F-box/WD-repeat proteins Pop1 and Pop2 form hetero- and homo-complexes together with cullin-1 in the fission yeast SCF (Skp1-Cullin-1-F-box) ubiquitin ligase.Genes Cells. 1998 Nov;3(11):721-35. doi: 10.1046/j.1365-2443.1998.00225.x. Genes Cells. 1998. PMID: 9990507
-
NEDD8 recruits E2-ubiquitin to SCF E3 ligase.EMBO J. 2001 Aug 1;20(15):4003-12. doi: 10.1093/emboj/20.15.4003. EMBO J. 2001. PMID: 11483504 Free PMC article.
-
Two distinct ubiquitin-proteolysis pathways in the fission yeast cell cycle.Philos Trans R Soc Lond B Biol Sci. 1999 Sep 29;354(1389):1551-7. doi: 10.1098/rstb.1999.0498. Philos Trans R Soc Lond B Biol Sci. 1999. PMID: 10582240 Free PMC article. Review.
-
Nedd8 modification of cul-1 activates SCF(beta(TrCP))-dependent ubiquitination of IkappaBalpha.Mol Cell Biol. 2000 Apr;20(7):2326-33. doi: 10.1128/MCB.20.7.2326-2333.2000. Mol Cell Biol. 2000. PMID: 10713156 Free PMC article.
-
F-box proteins and protein degradation: an emerging theme in cellular regulation.Plant Mol Biol. 2000 Sep;44(2):123-8. doi: 10.1023/a:1006413007456. Plant Mol Biol. 2000. PMID: 11117256 Review.
Cited by
-
Nedd8 processing enzymes in Schizosaccharomyces pombe.BMC Biochem. 2013 Mar 15;14:8. doi: 10.1186/1471-2091-14-8. BMC Biochem. 2013. PMID: 23496905 Free PMC article.
-
Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms.Genes Dev. 2003 May 1;17(9):1130-40. doi: 10.1101/gad.1090803. Epub 2003 Apr 14. Genes Dev. 2003. PMID: 12695334 Free PMC article.
-
Deletion mutants in COP9/signalosome subunits in fission yeast Schizosaccharomyces pombe display distinct phenotypes.Mol Biol Cell. 2002 Feb;13(2):493-502. doi: 10.1091/mbc.01-10-0521. Mol Biol Cell. 2002. PMID: 11854407 Free PMC article.
-
CRL4Mahj E3 ubiquitin ligase promotes neural stem cell reactivation.PLoS Biol. 2019 Jun 6;17(6):e3000276. doi: 10.1371/journal.pbio.3000276. eCollection 2019 Jun. PLoS Biol. 2019. PMID: 31170139 Free PMC article.
-
A new ubiquitin ligase involved in p57KIP2 proteolysis regulates osteoblast cell differentiation.EMBO Rep. 2008 Sep;9(9):878-84. doi: 10.1038/embor.2008.125. Epub 2008 Jul 25. EMBO Rep. 2008. PMID: 18660753 Free PMC article.
References
-
- Baumeister W., Walz,J., Zühl,F. and Seemüller,E. (1998) The proteasome: paradigm of a self-compartmentalizing protease. Cell, 92, 367–380. - PubMed
-
- Bochtler M., Ditzel,L., Groll,M., Hartmann,C. and Huber,H. (1999) The proteasome. Annu. Rev. Biophys. Biomol. Struct., 28, 295–317. - PubMed
-
- Carrano A., Eytan,E., Hershko,A. and Pagano,M. (1999) Skp2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nature Cell Biol., 1, 193–199. - PubMed
-
- Chen Y., McPhie,D.L., Hirschberg,J. and Neve,R.L. (2000) The amyloid precursor protein-binding protein APP-BP1 drives the cell cycle through the S–M checkpoint and causes apoptosis in neurons. J. Biol. Chem., 275, 8929–8935. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

