PathMaster: content-based cell image retrieval using automated feature extraction
- PMID: 10887168
- PMCID: PMC61444
- DOI: 10.1136/jamia.2000.0070404
PathMaster: content-based cell image retrieval using automated feature extraction
Abstract
Objective: Currently, when cytopathology images are archived, they are typically stored with a limited text-based description of their content. Such a description inherently fails to quantify the properties of an image and refers to an extremely small fraction of its information content. This paper describes a method for automatically indexing images of individual cells and their associated diagnoses by computationally derived cell descriptors. This methodology may serve to better index data contained in digital image databases, thereby enabling cytologists and pathologists to cross-reference cells of unknown etiology or nature.
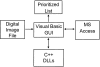
Design: The indexing method, implemented in a program called PathMaster, uses a series of computer-based feature extraction routines. Descriptors of individual cell characteristics generated by these routines are employed as indexes of cell morphology, texture, color, and spatial orientation.
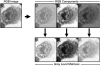
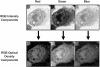
Measurements: The indexing fidelity of the program was tested after populating its database with images of 152 lymphocytes/lymphoma cells captured from lymph node touch preparations stained with hematoxylin and eosin. Images of "unknown" lymphoid cells, previously unprocessed, were then submitted for feature extraction and diagnostic cross-referencing analysis.
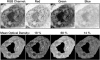
Results: PathMaster listed the correct diagnosis as its first differential in 94 percent of recognition trials. In the remaining 6 percent of trials, PathMaster listed the correct diagnosis within the first three "differentials."
Conclusion: PathMaster is a pilot cell image indexing program/search engine that creates an indexed reference of images. Use of such a reference may provide assistance in the diagnostic/prognostic process by furnishing a prioritized list of possible identifications for a cell of uncertain etiology.
Figures


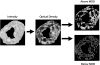

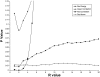


References
-
- O'Brien MJ. Sotnikov AV. Digital imaging in anatomic pathology [review]. Am J Clin Pathol. 1996;106(4 suppl 1): S25-32. - PubMed
-
- Niblack W. Query by image and video content: the QBIC system. IEEE Comput. 1995;28(9): 23-32.
-
- Wetzel AW, Andrews PL, Becich MJ, Gilbertson J. Computational aspects of pathology image classification and retrieval. J Supercomput. 1997;11: 279-93.
-
- Pentland A, Picard RW, Sclaroff S. Photobook: content-based manipulation of image databases. Int J Comput Vision. 1996;18(3): 233-54.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

