AtVPS45 complex formation at the trans-Golgi network
- PMID: 10888666
- PMCID: PMC14917
- DOI: 10.1091/mbc.11.7.2251
AtVPS45 complex formation at the trans-Golgi network
Abstract
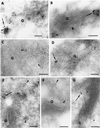
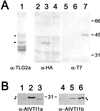
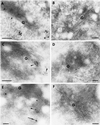
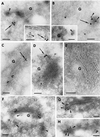
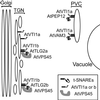
The Sec1p family of proteins are thought to be involved in the regulation of vesicle fusion reactions through interaction with t-SNAREs (target soluble N-ethylmaleimide-sensitive factor attachment protein receptors) at the target membrane. AtVPS45 is a member of this family from Arabidopsis thaliana that we now demonstrate to be present on the trans-Golgi network (TGN), where it colocalizes with the vacuolar cargo receptor AtELP. Unlike yeast Vps45p, AtVPS45 does not interact with, or colocalize with, the prevacuolar t-SNARE AtPEP12. Instead, AtVPS45 interacts with two t-SNAREs, AtTLG2a and AtTLG2b, that show similarity to the yeast t-SNARE Tlg2p. AtTLG2a and -b each colocalize with AtVPS45 at the TGN; however, AtTLG2a is in a different region of the TGN than AtTLG2b by immunogold electron microscopy. Therefore, we propose that complexes containing AtVPS45 and either AtTLG2a or -b define functional subdomains of the TGN and may be required for different trafficking events. Among other Arabidopsis SNAREs, AtVPS45 antibodies preferentially coprecipitate AtVTI1b over the closely related isoform AtVTI1a, implying that AtVTI1a and AtVTI1b also have distinct functions within the cell. These data point to a functional complexity within the plant secretory pathway, where proteins encoded by gene families have specialized functions, rather than functional redundancy.
Figures

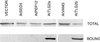


References
-
- Abeliovich H, Grote E, Novick P, Ferro-Novick S. Tlg2p, a yeast syntaxin homolog that resides on the Golgi and endocytic structures. J Biol Chem. 1998;273:11719–11727. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

