The Kar3p kinesin-related protein forms a novel heterodimeric structure with its associated protein Cik1p
- PMID: 10888675
- PMCID: PMC14926
- DOI: 10.1091/mbc.11.7.2373
The Kar3p kinesin-related protein forms a novel heterodimeric structure with its associated protein Cik1p
Abstract
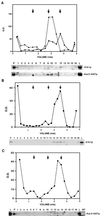
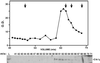
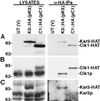
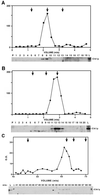
Proteins that physically associate with members of the kinesin superfamily are critical for the functional diversity observed for these microtubule motor proteins. However, quaternary structures of complexes between kinesins and kinesin-associated proteins are poorly defined. We have analyzed the nature of the interaction between the Kar3 motor protein, a minus-end-directed kinesin from yeast, and its associated protein Cik1. Extraction experiments demonstrate that Kar3p and Cik1p are tightly associated. Mapping of the interaction domains of the two proteins by two-hybrid analyses indicates that Kar3p and Cik1p associate in a highly specific manner along the lengths of their respective coiled-coil domains. Sucrose gradient velocity centrifugation and gel filtration experiments were used to determine the size of the Kar3-Cik1 complex from both mating pheromone-treated cells and vegetatively growing cells. These experiments predict a size for this complex that is consistent with that of a heterodimer containing one Kar3p subunit and one Cik1p subunit. Finally, immunoprecipitation of epitope-tagged and untagged proteins confirms that only one subunit of Kar3p and Cik1p are present in the Kar3-Cik1 complex. These findings demonstrate that the Kar3-Cik1 complex has a novel heterodimeric structure not observed previously for kinesin complexes.
Figures



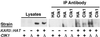

References
-
- Arndt K, Pelletier J, Muller K, Alber T, Michnick S, Pluckthun A. A heterodimeric coiled-coil peptide pair selected in vivo from a designed library-versus-library ensemble. J Mol Biol. 2000;295:627–639. - PubMed
-
- Berliner E, Young E, Anderson K, Mahtani H, Gelles J. Failure of single-headed kinesin to track parallel to microtubule protofilaments. Nature. 1995;373:718–721. - PubMed
-
- Bloom GS, Wagner MC, Pfister KK, Brady ST. Native structure and physical properties of bovine brain kinesin and identification of the ATP-binding subunit polypeptide. Biochemistry. 1988;27:3409–3416. - PubMed
-
- Brady S. A novel brain ATPase with properties expected for the fast axonal transport motor. Nature. 1985;317:73–75. - PubMed
-
- Brent R, Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985;43:729–736. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

