Complementary expression of two plastid-localized sigma-like factors in maize
- PMID: 10889237
- PMCID: PMC59051
- DOI: 10.1104/pp.123.3.883
Complementary expression of two plastid-localized sigma-like factors in maize
Abstract
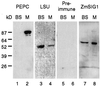
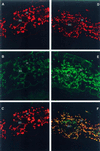
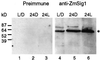
The eubacterial-like RNA polymerase of plastids is composed of organelle-encoded core subunits and nuclear-encoded sigma-factors. Families of sigma-like factors (SLFs) have been identified in several plants, including maize (Zea mays) and Arabidopsis. In vitro import assays determined that at least two of the maize sigma-like proteins have functional chloroplast transit peptides and thus are likely candidates for chloroplast transcriptional regulators. However, the roles of individual SLFs in chloroplast transcription remain to be determined. We have raised antibodies against the unique amino-terminal domains of two maize SLFs, ZmSig1 and ZmSig3, and have used these specific probes to examine the accumulation of each protein in different maize tissues and during chloroplast development. The expression of ZmSig1 is tissue specific and parallels the light-activated chloroplast development program in maize seedling leaves. Its accumulation in mature chloroplasts however, is not affected by subsequent changes in the light regime. It is interesting that the expression profile of ZmSig3 is complementary to that of ZmSig1. It accumulates in non-green tissues, including roots, etiolated seedling leaves, and the basal region of greening seedling leaves. The nonoverlapping expression patterns of these two plastid-localized SLFs suggest that they may direct differential expression of plastid genes during chloroplast development.
Figures


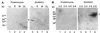


Similar articles
-
A nucleus-encoded maize protein with sigma factor activity accumulates in mitochondria and chloroplasts.Plant J. 2002 Jul;31(2):199-209. doi: 10.1046/j.1365-313x.2002.01344.x. Plant J. 2002. PMID: 12121449
-
Tissue-specific and light-dependent expression within a family of nuclear-encoded sigma-like factors from Zea mays.Mol Cell Biol Res Commun. 1999 Apr;1(1):14-20. doi: 10.1006/mcbr.1999.0102. Mol Cell Biol Res Commun. 1999. PMID: 10329472
-
Characterization of two chloroplast RNA polymerase sigma factors from Zea mays: photoregulation and differential expression.Proc Natl Acad Sci U S A. 1999 Apr 27;96(9):5316-21. doi: 10.1073/pnas.96.9.5316. Proc Natl Acad Sci U S A. 1999. PMID: 10220463 Free PMC article.
-
Role and regulation of plastid sigma factors and their functional interactors during chloroplast transcription - recent lessons from Arabidopsis thaliana.Eur J Cell Biol. 2010 Dec;89(12):940-6. doi: 10.1016/j.ejcb.2010.06.016. Epub 2010 Aug 10. Eur J Cell Biol. 2010. PMID: 20701995 Review.
-
The role of sigma factors in plastid transcription.Biochimie. 2000 Jun-Jul;82(6-7):537-48. doi: 10.1016/s0300-9084(00)00611-8. Biochimie. 2000. PMID: 10946105 Review.
Cited by
-
Chlamydomonas reinhardtii encodes a single sigma70-like factor which likely functions in chloroplast transcription.Curr Genet. 2006 May;49(5):333-40. doi: 10.1007/s00294-006-0060-7. Epub 2006 Feb 2. Curr Genet. 2006. PMID: 16453112
-
Characterization of Arabidopsis plastid sigma-like transcription factors SIG1, SIG2 and SIG3.Plant Mol Biol. 2003 Feb;51(3):385-99. doi: 10.1023/a:1022095017355. Plant Mol Biol. 2003. PMID: 12602869
-
Unique architecture of the plastid ribosomal RNA operon promoter recognized by the multisubunit RNA polymerase in tobacco and other higher plants.Plant Cell. 2003 Jan;15(1):195-205. doi: 10.1105/tpc.007914. Plant Cell. 2003. PMID: 12509531 Free PMC article.
-
Molecular genetic analysis of chloroplast gene promoters dependent on SIG2, a nucleus-encoded sigma factor for the plastid-encoded RNA polymerase, in Arabidopsis thaliana.Nucleic Acids Res. 2003 Dec 15;31(24):7090-8. doi: 10.1093/nar/gkg935. Nucleic Acids Res. 2003. PMID: 14654684 Free PMC article.
-
SIG1, a sigma factor for the chloroplast RNA polymerase, differently associates with multiple DNA regions in the chloroplast chromosomes in vivo.Int J Mol Sci. 2012 Sep 25;13(10):12182-94. doi: 10.3390/ijms131012182. Int J Mol Sci. 2012. PMID: 23202891 Free PMC article.
References
-
- Allison LA (2000) The role of ς factors in plastid transcription. Biochimie (in press) - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

