Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-beta and wnt pathways
- PMID: 10890911
- PMCID: PMC26952
- DOI: 10.1073/pnas.150152697
Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-beta and wnt pathways
Abstract
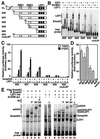
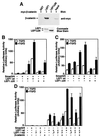
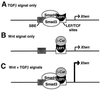
The transforming growth factor-beta (TGFbeta) and Wnt/wingless pathways play pivotal roles in tissue specification during development. Activation of Smads, the effectors of TGFbeta superfamily signals, results in Smad translocation from the cytoplasm into the nucleus where they act as transcriptional comodulators to regulate target gene expression. Wnt/wingless signals are mediated by the DNA-binding HMG box transcription factors lymphoid enhancer binding factor 1/T cell-specific factor (LEF1/TCF) and their coactivator beta-catenin. Herein, we show that Smad3 physically interacts with the HMG box domain of LEF1 and that TGFbeta and Wnt pathways synergize to activate transcription of the Xenopus homeobox gene twin (Xtwn). Disruption of specific Smad and LEF1/TCF DNA-binding sites in the promoter abrogates synergistic activation of the promoter. Consistent with this observation, introduction of Smad sites into a TGFbeta-insensitive LEF1/TCF target gene confers cooperative TGFbeta and Wnt responsiveness to the promoter. Furthermore, we demonstrate that TGFbeta-dependent activation of LEF1/TCF target genes can occur in the absence of beta-catenin binding to LEF1/TCF and requires both Smad and LEF1/TCF DNA-binding sites in the Xtwn promoter. Thus, our results show that TGFbeta and Wnt signaling pathways can independently or cooperatively regulate LEF1/TCF target genes and suggest a model for how these pathways can synergistically activate target genes.
Figures

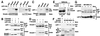


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

