Saccharomyces cerevisiae sigma 1278b has novel genes of the N-acetyltransferase gene superfamily required for L-proline analogue resistance
- PMID: 10894734
- PMCID: PMC101931
- DOI: 10.1128/JB.182.15.4249-4256.2000
Saccharomyces cerevisiae sigma 1278b has novel genes of the N-acetyltransferase gene superfamily required for L-proline analogue resistance
Abstract
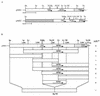
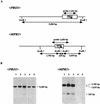
We discovered on the chromosome of Saccharomyces cerevisiae Sigma 1278b novel genes involved in L-proline analogue L-azetidine-2-carboxylic acid resistance which are not present in the standard laboratory strains. The 5.4 kb-DNA fragment was cloned from the genomic library of the L-azetidine-2-carboxylic acid-resistant mutant derived from a cross between S. cerevisiae strains S288C and Sigma 1278b. The nucleotide sequence of a 4.5-kb segment exhibited no identity with the sequence in the genome project involving strain S288C. Deletion analysis indicated that one open reading frame encoding a predicted protein of 229 amino acids is indispensable for L-azetidine-2-carboxylic acid resistance. The protein sequence was found to be a member of the N-acetyltransferase superfamily. Genomic Southern analysis and gene disruption showed that two copies of the novel gene with one amino acid change at position 85 required for L-azetidine-2-carboxylic acid resistance were present on chromosomes X and XIV of Sigma 1278b background strains. When this novel MPR1 or MPR2 gene (sigma 1278b gene for L-proline analogue resistance) was introduced into the other S. cerevisiae strains, all of the recombinants were resistant to L-azetidine-2-carboxylic acid, indicating that both MPR1 and MPR2 are expressed and have a global function in S. cerevisiae.
Figures




Similar articles
-
Polymorphism of the MPR1 gene required for toxic proline analogue resistance in the Saccharomyces cerevisiae complex species.Yeast. 2002 Dec;19(16):1437-45. doi: 10.1002/yea.927. Yeast. 2002. PMID: 12478591
-
Characterization of novel acetyltransferases found in budding and fission yeasts that detoxify a proline analogue, azetidine-2-carboxylic acid.J Biochem. 2003 Jan;133(1):67-74. doi: 10.1093/jb/mvg003. J Biochem. 2003. PMID: 12761200
-
A novel acetyltransferase found in Saccharomyces cerevisiae Sigma1278b that detoxifies a proline analogue, azetidine-2-carboxylic acid.J Biol Chem. 2001 Nov 9;276(45):41998-2002. doi: 10.1074/jbc.C100487200. Epub 2001 Sep 12. J Biol Chem. 2001. PMID: 11555637
-
MPR1 as a novel selection marker in Saccharomyces cerevisiae.Yeast. 2009 Nov;26(11):587-93. doi: 10.1002/yea.1708. Yeast. 2009. PMID: 19750564
-
A comprehensive review of the proline mimic azetidine-2-carboxylic acid (A2C).Toxicology. 2025 Jan;510:153999. doi: 10.1016/j.tox.2024.153999. Epub 2024 Nov 15. Toxicology. 2025. PMID: 39549916 Review.
Cited by
-
Genome Sequence and Analysis of a Stress-Tolerant, Wild-Derived Strain of Saccharomyces cerevisiae Used in Biofuels Research.G3 (Bethesda). 2016 Jun 1;6(6):1757-66. doi: 10.1534/g3.116.029389. G3 (Bethesda). 2016. PMID: 27172212 Free PMC article.
-
Selecting representative model micro-organisms.BMC Microbiol. 2005 May 17;5:26. doi: 10.1186/1471-2180-5-26. BMC Microbiol. 2005. PMID: 15904495 Free PMC article.
-
Genomic insights into the Saccharomyces sensu stricto complex.Genetics. 2015 Feb;199(2):281-91. doi: 10.1534/genetics.114.173633. Genetics. 2015. PMID: 25657346 Free PMC article. Review.
-
A nonconserved Ala401 in the yeast Rsp5 ubiquitin ligase is involved in degradation of Gap1 permease and stress-induced abnormal proteins.Proc Natl Acad Sci U S A. 2003 Sep 30;100(20):11505-10. doi: 10.1073/pnas.1933153100. Epub 2003 Sep 18. Proc Natl Acad Sci U S A. 2003. PMID: 14500784 Free PMC article.
-
Whole-genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae.PLoS Genet. 2011 Feb 3;7(2):e1001287. doi: 10.1371/journal.pgen.1001287. PLoS Genet. 2011. PMID: 21304888 Free PMC article.
References
-
- Altschul S F, Boguski M S, Gish W, Wooten J C. Issues in searching molecular sequence databases. Nat Genet. 1994;6:119–129. - PubMed
-
- Bonhivers M, Carbrey J M, Gould S J, Agre P. Aquaporins in Saccharomyces. Genetic and functional distinctions between laboratory and wild-type strains. J Biol Chem. 1998;273:27565–27572. - PubMed
-
- Breathnach R, Chambon P. Organization and expression of eukaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. - PubMed
-
- Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted and intracellular forms of yeast invertase. Cell. 1982;28:145–154. - PubMed
-
- Csonka L N. Proline over-production results in enhanced osmotolerance in Salmonella typhimurium. Mol Gen Genet. 1981;182:82–86. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

