A redox-responsive regulator of photosynthesis gene expression in the cyanobacterium Synechocystis sp. Strain PCC 6803
- PMID: 10894737
- PMCID: PMC101939
- DOI: 10.1128/JB.182.15.4268-4277.2000
A redox-responsive regulator of photosynthesis gene expression in the cyanobacterium Synechocystis sp. Strain PCC 6803
Abstract
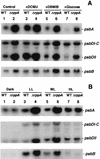
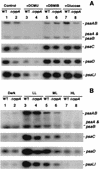
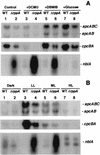
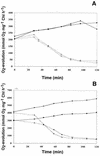
We have identified genes in the unicellular cyanobacterium Synechocystis sp. strain PCC 6803 that are involved with redox control of photosynthesis and pigment-related genes. The genes, rppA (sll0797) and rppB (sll0798), represent a two-component regulatory system that controls the synthesis of photosystem II (PSII) and PSI genes, in addition to photopigment-related genes. rppA (regulator of photosynthesis- and photopigment-related gene expression) and rppB exhibit strong sequence similarity to prokaryotic response regulators and histidine kinases, respectively. In the wild type, the steady-state mRNA levels of PSII reaction center genes increased when the plastoquinone (PQ) pool was oxidized and decreased when the PQ pool was reduced, whereas transcription of the PSI reaction center genes was affected in an opposite fashion. Such results suggested that the redox poise of the PQ pool is critical for regulation of the photosystem reaction center genes. In Delta rppA, an insertion mutation of rppA, the PSII gene transcripts were highly up-regulated relative to the wild type under all redox conditions, whereas transcription of phycobilisome-related genes and PSI genes was decreased. The higher transcription of the psbA gene in Delta rppA was manifest by higher translation of the D1 protein and a concomitant increase in O(2) evolution. The results demonstrated that RppA is a regulator of photosynthesis- and photopigment-related gene expression, is involved in the establishment of the appropriate stoichiometry between the photosystems, and can sense changes in the PQ redox poise.
Figures



References
-
- Alfonso M, Perewoska I, Constant S, Kirilovsky D. Redox control of psbA expression in cyanobacteria Synechocystis strains. J Photochem Photobiol B: Biol. 1999;48:104–113.
-
- Allen J F. Thylakoid protein phosphorylation, state 1-state 2 transitions, and photosystem stoichiometry adjustment: redox control at multiple levels of gene expression. Physiol Plant. 1995;93:196–205.
-
- Aro E-M, Virgin I, Andersson B. Photoinhibition of photosystem II: inactivation, protein damage and turnover. Biochim Biophys Acta. 1993;1143:113–134. - PubMed
-
- Barber J. Photosynthetic reaction centers: a common link. Trends Biochem Sci. 1987;12:321–326.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

