Identification, evolution, and essentiality of the mevalonate pathway for isopentenyl diphosphate biosynthesis in gram-positive cocci
- PMID: 10894743
- PMCID: PMC101949
- DOI: 10.1128/JB.182.15.4319-4327.2000
Identification, evolution, and essentiality of the mevalonate pathway for isopentenyl diphosphate biosynthesis in gram-positive cocci
Abstract
The mevalonate pathway and the glyceraldehyde 3-phosphate (GAP)-pyruvate pathway are alternative routes for the biosynthesis of the central isoprenoid precursor, isopentenyl diphosphate. Genomic analysis revealed that the staphylococci, streptococci, and enterococci possess genes predicted to encode all of the enzymes of the mevalonate pathway and not the GAP-pyruvate pathway, unlike Bacillus subtilis and most gram-negative bacteria studied, which possess only components of the latter pathway. Phylogenetic and comparative genome analyses suggest that the genes for mevalonate biosynthesis in gram-positive cocci, which are highly divergent from those of mammals, were horizontally transferred from a primitive eukaryotic cell. Enterococci uniquely encode a bifunctional protein predicted to possess both 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase and acetyl-CoA acetyltransferase activities. Genetic disruption experiments have shown that five genes encoding proteins involved in this pathway (HMG-CoA synthase, HMG-CoA reductase, mevalonate kinase, phosphomevalonate kinase, and mevalonate diphosphate decarboxylase) are essential for the in vitro growth of Streptococcus pneumoniae under standard conditions. Allelic replacement of the HMG-CoA synthase gene rendered the organism auxotrophic for mevalonate and severely attenuated in a murine respiratory tract infection model. The mevalonate pathway thus represents a potential antibacterial target in the low-G+C gram-positive cocci.
Figures
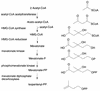




References
-
- Alberts A W, Chen J, Kuron G, Hunt V, Huff J, Hoffman C, Rothrock J, Lopez M, Joshua H, Harris E, Patchett A, Monaghan R, Currie S, Stapley E, Albers-Schonberg G, Hensens O, Hirshfield J, Hoogsteen K, Liesch J, Springer J. Mevinolin: a highly potent competitive inhibitor of hydroxymethylglutaryl-coenzyme A reductase and a cholesterol-lowering agent. Proc Natl Acad Sci USA. 1980;77:3957–3961. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

