A dual role for zinc fingers in both DNA binding and zinc sensing by the Zap1 transcriptional activator
- PMID: 10899124
- PMCID: PMC313982
- DOI: 10.1093/emboj/19.14.3704
A dual role for zinc fingers in both DNA binding and zinc sensing by the Zap1 transcriptional activator
Abstract
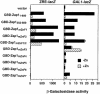
The Zap1 transcriptional activator of Saccharomyces cerevisiae controls zinc homeostasis. Zap1 induces target gene expression in zinc-limited cells and is repressed by high zinc. One such target gene is ZAP1 itself. In this report, we examine how zinc regulates Zap1 function. First, we show that transcriptional autoregulation of Zap1 is a minor component of zinc responsiveness; most regulation of Zap1 activity occurs post-translationally. Secondly, nuclear localization of Zap1 does not change in response to zinc, suggesting that zinc regulates DNA binding and/or activation domain function. To understand how Zap1 responds to zinc, we performed a functional dissection of the protein. Zap1 contains two activation domains. DNA-binding activity is conferred by five C-terminal C(2)H(2) zinc fingers and each finger is required for high-affinity DNA binding. The zinc-responsive domain of Zap1 also maps to the C-terminal zinc fingers. Furthermore, mutations that disrupt some of these fingers cause constitutive activity of a bifunctional Gal4 DNA-binding domain-Zap1 fusion protein. These results demonstrate a novel function of Zap1 zinc fingers in zinc sensing as well as DNA binding.
Figures



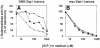


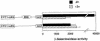
References
-
- Baeuerle P.A. and Baltimore,D. (1988) IκB: a specific inhibitor of the NF-κB transcription factor. Science, 242, 540–545. - PubMed
-
- Bird A.J., Evans-Galea,M., Blankman,E., Zhao,H., Luo,H., Winge,D.R. and Eide,D.J. (2000) Mapping the DNA binding domain of the Zap1 zinc-responsive transcriptional activator. J. Biol. Chem., 275, 16160–16166. - PubMed
-
- Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. - PubMed
-
- Cross F.R. (1997) ‘Marker swap’ plasmids: convenient tools for budding yeast molecular genetics. Yeast, 13, 647–663. - PubMed
-
- Cullin C. and Minvielle-Sebastia,L. (1994) Multipurpose vectors designed for the fast generation of N- or C-terminal epitope-tagged proteins. Yeast, 10, 105–112. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

