Cyclophilin A and Ess1 interact with and regulate silencing by the Sin3-Rpd3 histone deacetylase
- PMID: 10899127
- PMCID: PMC313981
- DOI: 10.1093/emboj/19.14.3739
Cyclophilin A and Ess1 interact with and regulate silencing by the Sin3-Rpd3 histone deacetylase
Abstract
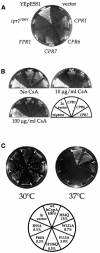
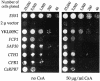
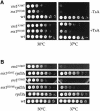
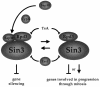
Three families of prolyl isomerases have been identified: cyclophilins, FK506-binding proteins (FKBPs) and parvulins. All 12 cyclophilins and FKBPs are dispensable for growth in yeast, whereas the one parvulin homolog, Ess1, is essential. We report here that cyclophilin A becomes essential when Ess1 function is compromised. We also show that overexpression of cyclophilin A suppresses ess1 conditional and null mutations, and that cyclophilin A enzymatic activity is required for suppression. These results indicate that cyclophilin A and Ess1 function in parallel pathways and act on common targets by a mechanism that requires prolyl isomerization. Using genetic and biochemical approaches, we found that one of these targets is the Sin3-Rpd3 histone deacetylase complex, and that cyclophilin A increases and Ess1 decreases disruption of gene silencing by this complex. We show that conditions that favor acetylation over deacetylation suppress ess1 mutations. Our findings support a model in which Ess1 and cyclophilin A modulate the activity of the Sin3-Rpd3 complex, and excess histone deacetylation causes mitotic arrest in ess1 mutants.
Figures





References
-
- Albert A., Lavoie,S. and Vincent,M. (1999) A hyperphosphorylated form of RNA polymerase II is the major interphase antigen of the phosphoprotein antibody MPM-2 and interacts with the peptidyl-prolyl isomerase Pin1. J. Cell Sci., 112, 2493–2500. - PubMed
-
- Anfinsen C.B. (1973) Principles that govern the folding of protein chains. Science, 181, 223–230. - PubMed
-
- Brillantes A.-M.B. et al. (1994) Stabilization of calcium release channel (ryanodine receptor) function by FK506-binding protein. Cell, 77, 513–523. - PubMed
-
- Bryk M., Banerjee,M., Murphy,M., Knudsen,K.E., Garfinkel,D.J. and Curcio,M.J. (1997) Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev., 11, 255–269. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

