SsrA-mediated tagging and proteolysis of LacI and its role in the regulation of lac operon
- PMID: 10899129
- PMCID: PMC313975
- DOI: 10.1093/emboj/19.14.3762
SsrA-mediated tagging and proteolysis of LacI and its role in the regulation of lac operon
Abstract
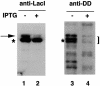
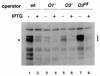
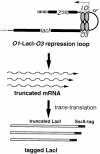
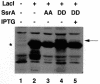
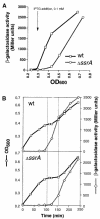
SsrA RNA of Escherichia coli, also known as 10Sa RNA or tmRNA, acts both as tRNA and mRNA when ribosomes are paused at the 3' end of an mRNA lacking a stop codon. This process, referred to as trans-translation, leads to the addition of a short peptide tag to the C-terminus of the incomplete nascent polypeptide. The tagged polypeptide is then degraded by C-terminal-specific proteases. Here, we focused on endogenous targets for the SsrA system and on a potential regulatory role of SsrA RNA. First, we show that trans-translation events occur frequently in normally growing E. COLI: cells. More specifically, we report that the lacI mRNA encoding Lac repressor (LacI) is a specific natural target for trans-translation. The binding of LacI to the lac operators results in truncated lacI mRNAs that are, in turn, recognized by the SsrA system. The SsrA-mediated tagging and proteolysis of LacI appears to play a role in cellular adaptation to lactose availability by supporting a rapid induction of lac operon expression.
Figures
References
-
- Aiba H., Adhya,S. and de Crombrugghe,B. (1981) Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem., 256, 11905–11910. - PubMed
-
- Atkins J.F. and Gesteland,R.F. (1996) A case for trans translation. Nature, 379, 769–771. - PubMed
-
- Casadaban M.J. (1976) Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage λ and µ. J. Mol. Biol., 104, 541–555. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

