m5C RNA and m5C DNA methyl transferases use different cysteine residues as catalysts
- PMID: 10899996
- PMCID: PMC26935
- DOI: 10.1073/pnas.97.15.8263
m5C RNA and m5C DNA methyl transferases use different cysteine residues as catalysts
Abstract
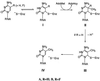
A family of RNA m(5)C methyl transferases (MTases) containing over 55 members in eight subfamilies has been identified recently by an iterative search of the genomic sequence databases by using the known 16S rRNA m(5)C 967 MTase, Fmu, as an initial probe. The RNA m(5)C MTase family contained sequence motifs that were highly homologous to motifs in the DNA m(5)C MTases, including the ProCys sequence that contains the essential Cys catalyst of the functionally similar DNA-modifying enzymes; it was reasonable to assign the Cys nucleophile to be that in the conserved ProCys. The family also contained an additional conserved Cys residue that aligns with the nucleophilic catalyst in m(5)U54 tRNA MTase. Surprisingly, the mutant of the putative Cys catalyst in the ProCys sequence was active and formed a covalent complex with 5-fluorocytosine-containing RNA, whereas the mutant at the other conserved Cys was inactive and unable to form the complex. Thus, notwithstanding the highly homologous sequences and similar functions, the RNA m(5)C MTase uses a different Cys as a catalytic nucleophile than the DNA m(5)C MTases. The catalytic Cys seems to be determined, not by the target base that is modified, but by whether the substrate is DNA or RNA. The function of the conserved ProCys sequence in the RNA m(5)C MTases remains unknown.
Figures


References
-
- Ivanetich K M, Santi D V. Prog Nucleic Acid Res Mol Biol. 1992;42:127–156. - PubMed
-
- Carreras C W, Santi D V. Annu Rev Biochem. 1995;64:721–762. - PubMed
-
- Gu X R, Gustafsson C, Ku J, Yu M, Santi D V. Biochemistry. 1999;38:4053–4057. - PubMed
-
- Cheng X. Annu Rev Biophys Biomol Struct. 1995;24:293–318. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

