The human cytomegalovirus 86-kilodalton major immediate-early protein interacts physically and functionally with histone acetyltransferase P/CAF
- PMID: 10906177
- PMCID: PMC112244
- DOI: 10.1128/jvi.74.16.7230-7237.2000
The human cytomegalovirus 86-kilodalton major immediate-early protein interacts physically and functionally with histone acetyltransferase P/CAF
Abstract
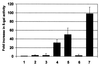
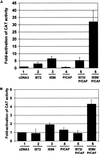
The major immediate-early proteins of human cytomegalovirus (HCMV) play a pivotal role in controlling viral and cellular gene expression during productive infection. As well as negatively autoregulating its own promoter, the HCMV 86-kDa major immediate early protein (IE86) activates viral early gene expression and is known to be a promiscuous transcriptional regulator of cellular genes. IE86 appears to act as a multimodal transcription factor. It is able to bind directly to target promoters to activate transcription but is also able to bridge between upstream binding factors such as CREB/ATF and the basal transcription complex as well as interacting directly with general transcription factors such as TATA-binding protein and TFIIB. We now show that IE86 is also able to interact directly with histone acetyltransferases during infection. At least one of these factors is the histone acetyltransferase CBP-associated factor (P/CAF). Furthermore, we show that this interaction results in synergistic transactivation by IE86 of IE86-responsive promoters. Recruitment of such chromatin-remodeling factors to target promoters by IE86 may help explain the ability of this viral protein to act as a promiscuous transactivator of cellular genes.
Figures



References
-
- Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. - PubMed
-
- Bannister A J, Oehler T, Wilhelm D, Angel P, Kouzarides T. Stimulation of c-Jun activity by CBP: c-Jun residues Ser63/73 are required for CBP induced stimulation in vivo and CBP binding in vitro. Oncogene. 1995;11:2509–2514. - PubMed
-
- Bresnahan W A, Albrecht T, Thompson E A. The cyclin E promoter is activated by human cytomegalovirus 86-kDa immediate early protein. J Biol Chem. 1998;273:22075–22082. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

