Common fold in helix-hairpin-helix proteins
- PMID: 10908318
- PMCID: PMC102670
- DOI: 10.1093/nar/28.14.2643
Common fold in helix-hairpin-helix proteins
Abstract
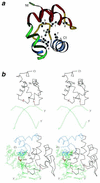
Helix-hairpin-helix (HhH) is a widespread motif involved in non-sequence-specific DNA binding. The majority of HhH motifs function as DNA-binding modules, however, some of them are used to mediate protein-protein interactions or have acquired enzymatic activity by incorporating catalytic residues (DNA glycosylases). From sequence and structural analysis of HhH-containing proteins we conclude that most HhH motifs are integrated as a part of a five-helical domain, termed (HhH)(2) domain here. It typically consists of two consecutive HhH motifs that are linked by a connector helix and displays pseudo-2-fold symmetry. (HhH)(2) domains show clear structural integrity and a conserved hydrophobic core composed of seven residues, one residue from each alpha-helix and each hairpin, and deserves recognition as a distinct protein fold. In addition to known HhH in the structures of RuvA, RadA, MutY and DNA-polymerases, we have detected new HhH motifs in sterile alpha motif and barrier-to-autointegration factor domains, the alpha-subunit of Escherichia coli RNA-polymerase, DNA-helicase PcrA and DNA glycosylases. Statistically significant sequence similarity of HhH motifs and pronounced structural conservation argue for homology between (HhH)(2) domains in different protein families. Our analysis helps to clarify how non-symmetric protein motifs bind to the double helix of DNA through the formation of a pseudo-2-fold symmetric (HhH)(2) functional unit.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

