PTIP, a novel BRCT domain-containing protein interacts with Pax2 and is associated with active chromatin
- PMID: 10908331
- PMCID: PMC102659
- DOI: 10.1093/nar/28.14.2741
PTIP, a novel BRCT domain-containing protein interacts with Pax2 and is associated with active chromatin
Abstract
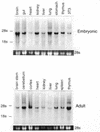
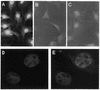
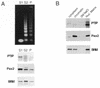
The Pax gene family encodes transcription factors essential for organ and tissue development in higher eukaryotes. Pax proteins are modular with an N-terminal DNA binding domain, a C-terminal transcription activation domain, and a transcription repression domain called the octapeptide. How these domains interact with the cellular machinery remains unclear. In this report, we describe the isolation and characterization of a novel gene and its encoded protein, PTIP, which binds to the activation domain of Pax2 and other Pax proteins. PTIP binds to Pax2 in vitro, in the yeast two-hybrid assay and in tissue culture cells. The binding of PTIP to Pax2 is inhibited by the octapeptide repression domain. The PTIP protein contains five BRCT domains, first identified in BRCA1 and other nuclear proteins involved in DNA repair/recombination or cell cycle control. Pax2 and PTIP co-localize in the cell nucleus to actively expressed chromatin and the nuclear matrix fraction. For the first time, these results point to a link between Pax transcription factors and active chromatin.
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

