RNA aptamers that specifically bind to a 16S ribosomal RNA decoding region construct
- PMID: 10908352
- PMCID: PMC102690
- DOI: 10.1093/nar/28.15.2902
RNA aptamers that specifically bind to a 16S ribosomal RNA decoding region construct
Abstract
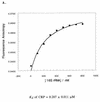
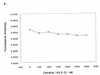
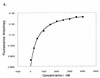
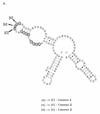
RNA-RNA recognition is a critical process in controlling many key biological events, such as translation and ribozyme functions. The recognition process governing RNA-RNA interactions can involve complementary Watson-Crick (WC) base pair binding, or can involve binding through tertiary structural interaction. Hence, it is of interest to determine which of the RNA-RNA binding events might emerge through an in vitro selection process. The A-site of the 16S rRNA decoding region was chosen as the target, both because it possesses several different RNA structural motifs, and because it is the rRNA site where codon/anticodon recognition occurs requiring recognition of both mRNA and tRNA. It is shown here that a single family of RNA molecules can be readily selected from two different sizes of RNA library. The tightest binding aptamer to the A-site 16S rRNA construct, 109.2-3, has its consensus sequences confined to a stem-loop region, which contains three nucleotides complementary to three of the four nucleotides in the stem-loop region of the A-site 16S rRNA. Point mutations on each of the three nucleotides on the stem-loop of the aptamer abolish its binding capacity. These studies suggest that the RNA aptamer 109.2-3 interacts with the simple 27 nt A-site decoding region of 16S rRNA through their respective stem-loops. The most probable mode of interaction is through complementary WC base pairing, commonly referred to as a loop-loop 'kissing' motif. High affinity binding to the other structural motifs in the decoding region were not observed.
Figures






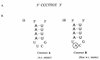


References
-
- Ellington A.D. and Szostak,J.W. (1990) Nature, 346, 818–822. - PubMed
-
- Tuerk C. and Gold,L. (1990) Science, 249, 505–510. - PubMed
-
- Beaudry A. and Joyce,G.F. (1992) Science, 257, 635–641. - PubMed
-
- Gold L., Polisky,B., Uhlenbeck,O. and Yarus,M. (1995) Annu. Rev. Biochem., 64, 763–797. - PubMed
-
- Pei D., Ulrich,H.D. and Schultz,P.G. (1991) Science, 253, 1408–1411. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

