Compaction of DNA in an anionic micelle environment followed by assembly into phosphatidylcholine liposomes
- PMID: 10908363
- PMCID: PMC102676
- DOI: 10.1093/nar/28.15.2986
Compaction of DNA in an anionic micelle environment followed by assembly into phosphatidylcholine liposomes
Abstract
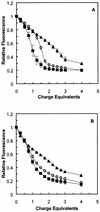
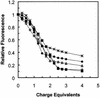
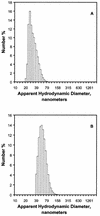
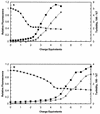
A difficult problem concerning the interaction of DNA with amphiphiles of opposite charge above their critical micelle concentration is the propensity for aggregation of the condensed DNA complexes. In this study, this problem was addressed by attenuating amphiphile charge density within a cholate micelle environment. The amphiphile consisted of a cationic peptide, acetyl-CWKKKPKK-amide, conjugated to dilaurylphos-phatidylethanolamine. In the presence of cholate, multiple equivalents of cationic charge were required to bring about the completion of DNA condensation. At the end point of condensation, stable, soluble DNA-micelle complexes were formed, which by dynamic light scattering exhibited apparent hydro-dynamic diameters between 30 and 60 nm. Aggregation, as measured by static light scattering at 90 degrees and by turbidity, was not observed until further additions of peptide-lipid conjugate were made beyond the end point of DNA condensation. Liposome complexes containing the non-aggregated, compacted DNA were formed by adding dioleoylphosphatidylcholine followed by removing the cholate by dialysis. The resulting complexes were distributed within a narrow density range, the DNA was quantitatively assembled into the liposomes, and liposomes without DNA were not detected. Small particles were formed with a mean hydrodynamic diameter of 77 nm. The liposomal DNA showed complete retention of its supercoiled form and no detectable sensitivity to DNase (25 U/10 microg DNA, 1.5 h, 37 degrees C). The use of an anionic, dialyzable amphiphile to attenuate charge inter-actions between DNA and cationic amphiphiles is a useful technology for the quantitative assembly of compacted DNA into conventional liposomes, with complete protection against nuclease activity.
Figures




References
-
- Fraley R. and Papahadjopoulos,D. (1982) Curr. Top. Microbiol. Immunol., 96, 171–191. - PubMed
-
- Lurquin P.F. (1993) In Gregoriadis,G. (ed.), Liposome Technology, Vol. II, 2nd Edn. CRC Press, Boca Raton, FL, pp. 129–139.
-
- Felgner P.L., Barenholz,Y., Behr,J.P., Cheng,S.H., Cullis,P., Huang,L., Jessee,J.A., Seymour,L., Szoka,F. and Thierry,A.R. (1997) Hum. Gene Ther., 8, 511–512. - PubMed
-
- Lasic D.D. (1997) Liposomes in Gene Delivery. CRC Press, Boca Raton, FL.

