Genes encoding Drosophila melanogaster RNA polymerase II general transcription factors: diversity in TFIIA and TFIID components contributes to gene-specific transcriptional regulation
- PMID: 10908585
- PMCID: PMC2180226
- DOI: 10.1083/jcb.150.2.f45
Genes encoding Drosophila melanogaster RNA polymerase II general transcription factors: diversity in TFIIA and TFIID components contributes to gene-specific transcriptional regulation
Figures
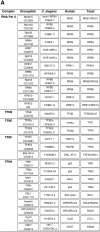

References
-
- Adams M.D., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F. The genome sequence of Drosophila melanogaster . Science. 2000;287:2185–2195. - PubMed
-
- Albright S.R., Tjian R. TAFs revisitedmore data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. - PubMed
-
- Bell D., Tora L. Regulation of gene expression by multiple forms of TFIID and other TAFII-containing complexes. Exp. Cell Res. 1999;246:11–19. - PubMed
-
- Birck C., Poch O., Romier C., Ruff M., Mengus G., Lavigne A.-C., Davidson I., Moras D. Human TAFII28 and TAFII18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell. 1998;94:239–249. - PubMed
-
- Brown C.E., Lechner T., Howe L., Workman J.L. The many hats of transcriptional coactivators. Trends Biochem. Sci. 2000;25:15–19. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

