The molecular basis of vancomycin resistance in clinically relevant Enterococci: crystal structure of D-alanyl-D-lactate ligase (VanA)
- PMID: 10908650
- PMCID: PMC16797
- DOI: 10.1073/pnas.150116497
The molecular basis of vancomycin resistance in clinically relevant Enterococci: crystal structure of D-alanyl-D-lactate ligase (VanA)
Abstract
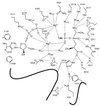
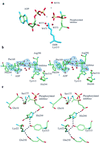
d-alanine-d-lactate ligase from Enterococcus faecium BM4147 is directly responsible for the biosynthesis of alternate cell-wall precursors in bacteria, which are resistant to the glycopeptide antibiotic vancomycin. The crystal structure has been determined with data extending to 2.5-A resolution. This structure shows that the active site has unexpected interactions and is distinct from previous models for d-alanyl-d-lactate ligase mechanistic studies. It appears that the preference of the enzyme for lactate as a ligand over d-alanine could be mediated by electrostatic effects and/or a hydrogen-bonding network, which principally involve His-244. The structure of d-alanyl-d-lactate ligase provides a revised interpretation of the molecular events that lead to vancomycin resistance.
Figures

References
-
- Novak R, Henriques B, Charpentier E, Normark S, Tuomanen E. Nature (London) 1999;399:580–593.
-
- Walsh C T, Fisher S L, Park I-S, Prahalad M, Wu Z. Chem Biol. 1996;3:21–28. - PubMed
-
- Bugg T D H, Wright G, Dutka-Malen S, Arthur M, Courvalin P, Walsh C T. Biochemistry. 1991;30:10408–10415. - PubMed
-
- Park I-S, Lin C-H, Walsh C T. Biochemistry. 1996;35:10464–10471. - PubMed
-
- Reynolds P E, Depardieu F, Dutka-Malen S, Arthur M, Courvalin P. Mol Microbiol. 1994;13:1065–1070. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

