Molecular identification of oligoalginate lyase of Sphingomonas sp. strain A1 as one of the enzymes required for complete depolymerization of alginate
- PMID: 10913091
- PMCID: PMC94629
- DOI: 10.1128/JB.182.16.4572-4577.2000
Molecular identification of oligoalginate lyase of Sphingomonas sp. strain A1 as one of the enzymes required for complete depolymerization of alginate
Abstract
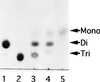
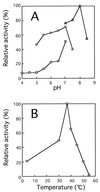
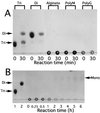
A bacterium, Sphingomonas sp. strain A1, can incorporate alginate into cells through a novel ABC (ATP-binding cassette) transporter system specific to the macromolecule. The transported alginate is depolymerized to di- and trisaccharides by three kinds of cytoplasmic alginate lyases (A1-I [66 kDa], A1-II [25 kDa], and A1-III [40 kDa]) generated from a single precursor through posttranslational autoprocessing. The resultant alginate oligosaccharides were degraded to monosaccharides by cytoplasmic oligoalginate lyase. The enzyme and its gene were isolated from the bacterial cells grown in the presence of alginate. The purified enzyme was a monomer with a molecular mass of 85 kDa and cleaved glycosidic bonds not only in oligosaccharides produced from alginate by alginate lyases but also in polysaccharides (alginate, polymannuronate, and polyguluronate) most efficiently at pH 8.0 and 37 degrees C. The reaction catalyzed by the oligoalginate lyase was exolytic and thought to play an important role in the complete depolymerization of alginate in Sphingomonas sp. strain A1. The gene for this novel enzyme consisted of an open reading frame of 2,286 bp encoding a polypeptide with a molecular weight of 86,543 and was located downstream of the genes coding for the precursor of alginate lyases (aly) and the ABC transporter (algS, algM1, and algM2). This result indicates that the genes for proteins required for the transport and complete depolymerization of alginate are assembled to form a cluster.
Figures



References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience, John Wiley and Sons; 1987.
-
- Boat T F, Beadet A L, Welsh M J. The metabolic basis of inherited disease. In: Seriver C R, editor. Cystic fibrosis. New York, N.Y: McGraw-Hill; 1989. pp. 2649–2680.
-
- Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
-
- Gacesa P. Alginates. Carbohydr Polym. 1988;8:161–182.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

