sigma(BldN), an extracytoplasmic function RNA polymerase sigma factor required for aerial mycelium formation in Streptomyces coelicolor A3(2)
- PMID: 10913095
- PMCID: PMC94633
- DOI: 10.1128/JB.182.16.4606-4616.2000
sigma(BldN), an extracytoplasmic function RNA polymerase sigma factor required for aerial mycelium formation in Streptomyces coelicolor A3(2)
Abstract
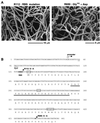
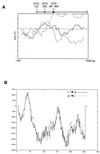
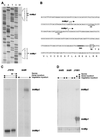
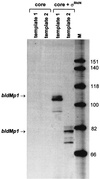
Sporulation mutants of Streptomyces coelicolor appear white because they are defective in the synthesis of the gray polyketide spore pigment, and such white (whi) mutants have been used to define 13 sporulation loci. whiN, one of five new whi loci identified in a recent screen of NTG (N-methyl-N'-nitro-N-nitrosoguanidine)-induced whi strains (N. J. Ryding et al., J. Bacteriol. 181:5419-5425, 1999), was defined by two mutants, R112 and R650. R650 produced frequent spores that were longer than those of the wild type. In contrast, R112 produced long, straight, undifferentiated hyphae, although rare spore chains were observed, sometimes showing highly irregular septum placement. Subcloning and sequencing showed that whiN encodes a member of the extracytoplasmic function subfamily of RNA polymerase sigma factors and that the sigma factor has an unusual N-terminal extension of approximately 86 residues that is not present in other sigma factors. A constructed whiN null mutant failed to form aerial mycelium (the "bald" phenotype) and, as a consequence, whiN was renamed bldN. This observation was not totally unexpected because, on some media, the R112 point mutant produced substantially less aerial mycelium than its parent, M145. The bldN null mutant did not fit simply into the extracellular signaling cascade proposed for S. coelicolor bld mutants. Expression of bldN was analyzed during colony development in wild-type and aerial mycelium-deficient bld strains. bldN was transcribed from a single promoter, bldNp. bldN transcription was developmentally regulated, commencing approximately at the time of aerial mycelium formation, and depended on bldG and bldH, but not on bldA, bldB, bldC, bldF, bldK, or bldJ or on bldN itself. Transcription from the p1 promoter of the response-regulator gene bldM depended on bldN in vivo, and the bldMp1 promoter was shown to be a direct biochemical target for sigma(BldN) holoenzyme in vitro.
Figures



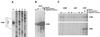
References
-
- Alper S, Dufour A, Garsin D A, Duncan L, Losick R. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J Mol Biol. 1996;260:165–177. - PubMed
-
- Bibb M J, Findlay P R, Johnson M W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984;30:157–166. - PubMed
-
- Bierman M, Logan R, O'Brien K, Seno E T, Rao R N, Schoner B E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. - PubMed
-
- Buttner M J, Fearnley I M, Bibb M J. The agarase gene (dagA) of Streptomyces coelicolor A3(2): nucleotide sequence and transcriptional analysis. Mol Gen Genet. 1987;209:101–109. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

