3-Deoxy-D-manno-oct-2-ulosonic acid (Kdo) transferase of Legionella pneumophila transfers two kdo residues to a structurally different lipid A precursor of Escherichia coli
- PMID: 10913104
- PMCID: PMC94642
- DOI: 10.1128/JB.182.16.4654-4657.2000
3-Deoxy-D-manno-oct-2-ulosonic acid (Kdo) transferase of Legionella pneumophila transfers two kdo residues to a structurally different lipid A precursor of Escherichia coli
Abstract
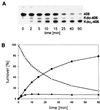
The 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo) transferase gene of Legionella pneumophila was cloned and sequenced. Despite remarkable structural differences in lipid A, the gene complemented a corresponding Escherichia coli mutant and was shown to encode a bifunctional enzyme which transferred 2 Kdo residues to a lipid A acceptor of E. coli.
Figures



Similar articles
-
Cloning and characterization of 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo) transferase genes (kdtA) from Acinetobacter baumannii and Acinetobacter haemolyticus.Eur J Biochem. 1998 Jun 1;254(2):404-12. doi: 10.1046/j.1432-1327.1998.2540404.x. Eur J Biochem. 1998. PMID: 9660198
-
3-Deoxy-D-manno-oct-2-ulosonic acid (Kdo) transferase (WaaA) and kdo kinase (KdkA) of Haemophilus influenzae are both required to complement a waaA knockout mutation of Escherichia coli.J Biol Chem. 2000 Nov 10;275(45):34954-62. doi: 10.1074/jbc.M005204200. J Biol Chem. 2000. PMID: 10952982
-
Biosynthesis of endotoxins. Purification and catalytic properties of 3-deoxy-D-manno-octulosonic acid transferase from Escherichia coli.J Biol Chem. 1992 May 15;267(14):9988-97. J Biol Chem. 1992. PMID: 1577828
-
Biochemical Characterization of Bifunctional 3-Deoxy-β-d-manno-oct-2-ulosonic Acid (β-Kdo) Transferase KpsC from Escherichia coli Involved in Capsule Biosynthesis.J Biol Chem. 2016 Oct 7;291(41):21519-21530. doi: 10.1074/jbc.M116.751115. Epub 2016 Aug 17. J Biol Chem. 2016. PMID: 27535220 Free PMC article.
-
Chlamydial lipopolysaccharide.Biochim Biophys Acta. 1999 Oct 8;1455(2-3):387-402. doi: 10.1016/s0925-4439(99)00061-7. Biochim Biophys Acta. 1999. PMID: 10571027 Review.
Cited by
-
Characterization of the gene encoding the major secreted lysophospholipase A of Legionella pneumophila and its role in detoxification of lysophosphatidylcholine.Infect Immun. 2002 Nov;70(11):6094-106. doi: 10.1128/IAI.70.11.6094-6106.2002. Infect Immun. 2002. PMID: 12379686 Free PMC article.
-
Endotoxin of Neisseria meningitidis composed only of intact lipid A: inactivation of the meningococcal 3-deoxy-D-manno-octulosonic acid transferase.J Bacteriol. 2002 May;184(9):2379-88. doi: 10.1128/JB.184.9.2379-2388.2002. J Bacteriol. 2002. PMID: 11948150 Free PMC article.
-
WaaA of the hyperthermophilic bacterium Aquifex aeolicus is a monofunctional 3-deoxy-D-manno-oct-2-ulosonic acid transferase involved in lipopolysaccharide biosynthesis.J Biol Chem. 2009 Aug 14;284(33):22248-22262. doi: 10.1074/jbc.M109.033308. Epub 2009 Jun 22. J Biol Chem. 2009. PMID: 19546212 Free PMC article.
-
Legionella dumoffii DjlA, a member of the DnaJ family, is required for intracellular growth.Infect Immun. 2004 Jun;72(6):3592-603. doi: 10.1128/IAI.72.6.3592-3603.2004. Infect Immun. 2004. PMID: 15155669 Free PMC article.
References
-
- Altschul S F, Gish W. Local alignment statistics. Methods Enzymol. 1996;266:460–480. - PubMed
-
- Belunis C J, Raetz C R H. Biosynthesis of endotoxin. Purification and catalytic properties of 3-deoxy-d-manno-octulosonic acid transferase from Escherichia coli. J Biol Chem. 1992;267:9988–9997. - PubMed
-
- Belunis C J, Clementz T, Carty S M, Raetz C R H. Inhibition of lipopolysaccharide biosynthesis and cell growth following inactivation of the kdtA gene in Escherichia coli. J Biol Chem. 1995;270:27646–27652. - PubMed
-
- Belunis C J, Mdluli K E, Raetz C R H, Nano F E. A novel 3-deoxy-d-manno-octulosonic acid transferase from Chlamydia trachomatis required for the expression of the genus-specific epitope. J Biol Chem. 1992;267:18702–18707. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

