Receptor-like protein tyrosine phosphatase alpha homodimerizes on the cell surface
- PMID: 10913175
- PMCID: PMC86069
- DOI: 10.1128/MCB.20.16.5917-5929.2000
Receptor-like protein tyrosine phosphatase alpha homodimerizes on the cell surface
Abstract
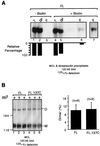
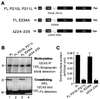
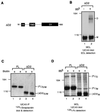
We reported previously that the N-terminal D1 catalytic domain of receptor protein-tyrosine phosphatase alpha (RPTPalpha) forms a symmetrical, inhibited dimer in a crystal structure, in which a helix-turn-helix wedge element from one monomer is inserted into the catalytic cleft of the other monomer. Previous functional studies also suggested that dimerization inhibits the biological activity of a CD45 chimeric RPTP and the catalytic activity of an isolated RPTPsigma D1 catalytic domain. Most recently, we have also shown that enforced dimerization inhibits the biological activity of full-length RPTPalpha in a wedge-dependent manner. The physiological significance of such inhibition is unknown, due to a lack of understanding of how RPTPalpha dimerization is regulated in vivo. In this study, we show that transiently expressed cell surface RPTPalpha exists predominantly as homodimers, suggesting that dimerization-mediated inhibition of RPTPalpha biological activity is likely to be physiologically relevant. Consistent with our published and unpublished crystallographic data, we show that mutations in the wedge region of D1 catalytic domain and deletion of the entire D2 catalytic domain independently reduced but did not abolish RPTPalpha homodimerization, suggesting that both domains are critically involved but that neither is essential for homodimerization. Finally, we also provide evidence that both the RPTPalpha extracellular domain and the transmembrane domain were independently able to homodimerize. These results lead us to propose a zipper model in which inactive RPTPalpha dimers are stabilized by multiple, relatively weak dimerization interfaces. Dimerization in this manner would provide a potential mechanism for negative regulation of RPTPalpha. Such RPTPalpha dimers could be activated by extracellular ligands or intracellular binding proteins that induce monomerization or by intracellular signaling events that induce an open conformation of the dimer.
Figures

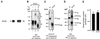



Similar articles
-
H2O2-induced intermolecular disulfide bond formation between receptor protein-tyrosine phosphatases.J Biol Chem. 2004 Oct 22;279(43):44355-61. doi: 10.1074/jbc.M407483200. Epub 2004 Aug 4. J Biol Chem. 2004. PMID: 15294898
-
Dimerization inhibits the activity of receptor-like protein-tyrosine phosphatase-alpha.Nature. 1999 Oct 7;401(6753):606-10. doi: 10.1038/44170. Nature. 1999. PMID: 10524630
-
RPTPα phosphatase activity is allosterically regulated by the membrane-distal catalytic domain.J Biol Chem. 2020 Apr 10;295(15):4923-4936. doi: 10.1074/jbc.RA119.011808. Epub 2020 Mar 5. J Biol Chem. 2020. PMID: 32139509 Free PMC article.
-
Meeting at mitosis: cell cycle-specific regulation of c-Src by RPTPalpha.Sci STKE. 2002 Jan 15;2002(115):pe3. doi: 10.1126/stke.2002.115.pe3. Sci STKE. 2002. PMID: 11796915 Review.
-
Receptor protein-tyrosine phosphatase signalling in development.Int J Dev Biol. 1999;43(7):723-33. Int J Dev Biol. 1999. PMID: 10668981 Review.
Cited by
-
RPTP-alpha acts as a transducer of mechanical force on alphav/beta3-integrin-cytoskeleton linkages.J Cell Biol. 2003 Apr 14;161(1):143-53. doi: 10.1083/jcb.200211061. Epub 2003 Apr 7. J Cell Biol. 2003. PMID: 12682088 Free PMC article.
-
The receptor protein tyrosine phosphatase LAR promotes R7 photoreceptor axon targeting by a phosphatase-independent signaling mechanism.Proc Natl Acad Sci U S A. 2009 Nov 17;106(46):19399-404. doi: 10.1073/pnas.0903961106. Epub 2009 Nov 4. Proc Natl Acad Sci U S A. 2009. PMID: 19889974 Free PMC article.
-
Aberrant DNA methylation of PTPRG as one possible mechanism of its under-expression in CML patients in the State of Qatar.Mol Genet Genomic Med. 2020 Oct;8(10):e1319. doi: 10.1002/mgg3.1319. Epub 2020 Jul 23. Mol Genet Genomic Med. 2020. PMID: 32700424 Free PMC article.
-
Inhibition of mitogen-activated protein kinase phosphatase 3 activity by interdomain binding.J Biol Chem. 2008 Oct 17;283(42):28574-83. doi: 10.1074/jbc.M801747200. Epub 2008 Aug 11. J Biol Chem. 2008. PMID: 18694935 Free PMC article.
-
Two mechanisms activate PTPalpha during mitosis.EMBO J. 2001 Nov 1;20(21):6037-49. doi: 10.1093/emboj/20.21.6037. EMBO J. 2001. PMID: 11689444 Free PMC article.
References
-
- Barnea G, Grumet M, Milev P, Silvennoinen O, Levy J B, Sap J, Schlessinger J. Receptor tyrosine phosphatase β is expressed in the form of proteoglycan and binds to the extracellular matrix protein tenascin. J Biol Chem. 1994;269:14349–14352. - PubMed
-
- Bhandari V, Lim K L, Pallen C J. Physical and functional interactions between receptor-like protein-tyrosine phosphatase α and p59fyn. J Biol Chem. 1998;273:8691–8698. - PubMed
-
- Bilwes A M, den Hertog J, Hunter T, Noel J P. Structural basis for inhibition of receptor protein-tyrosine phosphatase α by dimerization. Nature. 1996;382:555–559. - PubMed
-
- Buist, A., C. Blanchetot, L. G. Tertoolen, and J. den Hertog. Identification of p130Cas as an in vivo substrate of receptor protein-tyrosine phosphatase α. J. Biol. Chem., in press. - PubMed
-
- Buist A, Zhang Y L, Keng Y F, Wu L, Zhang Z Y, den Hertog J. Restoration of potent protein-tyrosine phosphatase activity into the membrane-distal domain of receptor protein-tyrosine phosphatase α. Biochemistry. 1999;38:914–922. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
