Rap2 as a slowly responding molecular switch in the Rap1 signaling cascade
- PMID: 10913189
- PMCID: PMC86083
- DOI: 10.1128/MCB.20.16.6074-6083.2000
Rap2 as a slowly responding molecular switch in the Rap1 signaling cascade
Abstract
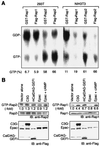
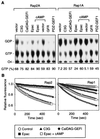
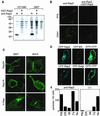
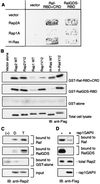
Rap2 is a member of the Ras family of GTPases and exhibits 60% identity to Rap1, but the function and regulation of Rap2 remain obscure. We found that, unlike the other Ras family proteins, the GTP-bound active form exceeded 50% of total Rap2 protein in adherent cells. Guanine nucleotide exchange factors (GEFs) for Rap1, C3G, Epac (or cyclic AMP [cAMP]-GEF), CalDAG-GEFI, PDZ-GEF1, and GFR efficiently increased the level of GTP-Rap2 both in 293T cells and in vitro. GTPase-activating proteins (GAPs) for Rap1, rap1GAPII and SPA-1, stimulated Rap2 GTPase, but with low efficiency. The half-life of GTP-Rap2 was significantly longer than that of GTP-Rap1 in 293T cells, indicating that low sensitivity to GAPs caused a high GTP/GDP ratio on Rap2. Rap2 bound to the Ras-binding domain of Raf and inhibited Ras-dependent activation of Elk1 transcription factor, as did Rap1. The level of GTP-Rap2 in rat 3Y1 fibroblasts was decreased by the expression of v-Src, and expression of a GTPase-deficient Rap2 mutant inhibited v-Src-dependent transformation of 3Y1 cells. Altogether, Rap2 is regulated by a similar set of GEFs and GAPs as Rap1 and functions as a slowly responding molecular switch in the Rap1 signaling cascade.
Figures





Similar articles
-
[Regulation of a small GTPase Rap2].Hokkaido Igaku Zasshi. 2001 Jan;76(1):13-20. Hokkaido Igaku Zasshi. 2001. PMID: 11235208 Japanese.
-
Identification of guanine nucleotide exchange factors (GEFs) for the Rap1 GTPase. Regulation of MR-GEF by M-Ras-GTP interaction.J Biol Chem. 2000 Nov 10;275(45):34901-8. doi: 10.1074/jbc.M005327200. J Biol Chem. 2000. PMID: 10934204
-
Characterisation of PDZ-GEFs, a family of guanine nucleotide exchange factors specific for Rap1 and Rap2.Biochim Biophys Acta. 2003 Feb 17;1593(2-3):141-9. doi: 10.1016/s0167-4889(02)00365-8. Biochim Biophys Acta. 2003. PMID: 12581858
-
Ras and Rap1: two highly related small GTPases with distinct function.Exp Cell Res. 1999 Nov 25;253(1):157-65. doi: 10.1006/excr.1999.4695. Exp Cell Res. 1999. PMID: 10579920 Review.
-
Rap-linked cAMP signaling Epac proteins: compartmentation, functioning and disease implications.Cell Signal. 2011 Aug;23(8):1257-66. doi: 10.1016/j.cellsig.2011.03.007. Epub 2011 Mar 22. Cell Signal. 2011. PMID: 21402149 Review.
Cited by
-
Complex interplay between RAS GTPases and RASSF effectors regulates subcellular localization of YAP.EMBO Rep. 2024 Aug;25(8):3574-3600. doi: 10.1038/s44319-024-00203-9. Epub 2024 Jul 15. EMBO Rep. 2024. PMID: 39009833 Free PMC article.
-
RAP1GAP inhibits cytoskeletal remodeling and motility in thyroid cancer cells.Endocr Relat Cancer. 2012 Jul 22;19(4):575-88. doi: 10.1530/ERC-12-0086. Print 2012 Aug. Endocr Relat Cancer. 2012. PMID: 22696507 Free PMC article.
-
The initial enzyme for glycosylphosphatidylinositol biosynthesis requires PIG-Y, a seventh component.Mol Biol Cell. 2005 Nov;16(11):5236-46. doi: 10.1091/mbc.e05-08-0743. Epub 2005 Sep 14. Mol Biol Cell. 2005. PMID: 16162815 Free PMC article.
-
TC-PTP dephosphorylates the guanine nucleotide exchange factor C3G (RapGEF1) and negatively regulates differentiation of human neuroblastoma cells.PLoS One. 2011;6(8):e23681. doi: 10.1371/journal.pone.0023681. Epub 2011 Aug 18. PLoS One. 2011. PMID: 21876762 Free PMC article.
-
GTPase activity of Di-Ras proteins is stimulated by Rap1GAP proteins.Small GTPases. 2010 Nov;1(3):133-141. doi: 10.4161/sgtp.1.3.14742. Small GTPases. 2010. PMID: 21686267 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
