Community structure, cellular rRNA content, and activity of sulfate-reducing bacteria in marine arctic sediments
- PMID: 10919825
- PMCID: PMC92189
- DOI: 10.1128/AEM.66.8.3592-3602.2000
Community structure, cellular rRNA content, and activity of sulfate-reducing bacteria in marine arctic sediments
Abstract
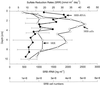
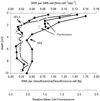
The community structure of sulfate-reducing bacteria (SRB) of a marine Arctic sediment (Smeerenburgfjorden, Svalbard) was characterized by both fluorescence in situ hybridization (FISH) and rRNA slot blot hybridization by using group- and genus-specific 16S rRNA-targeted oligonucleotide probes. The SRB community was dominated by members of the Desulfosarcina-Desulfococcus group. This group accounted for up to 73% of the SRB detected and up to 70% of the SRB rRNA detected. The predominance was shown to be a common feature for different stations along the coast of Svalbard. In a top-to-bottom approach we aimed to further resolve the composition of this large group of SRB by using probes for cultivated genera. While this approach failed, directed cloning of probe-targeted genes encoding 16S rRNA was successful and resulted in sequences which were all affiliated with the Desulfosarcina-Desulfococcus group. A group of clone sequences (group SVAL1) most closely related to Desulfosarcina variabilis (91.2% sequence similarity) was dominant and was shown to be most abundant in situ, accounting for up to 54. 8% of the total SRB detected. A comparison of the two methods used for quantification showed that FISH and rRNA slot blot hybridization gave comparable results. Furthermore, a combination of the two methods allowed us to calculate specific cellular rRNA contents with respect to localization in the sediment profile. The rRNA contents of Desulfosarcina-Desulfococcus cells were highest in the first 5 mm of the sediment (0.9 and 1.4 fg, respectively) and decreased steeply with depth, indicating that maximal metabolic activity occurred close to the surface. Based on SRB cell numbers, cellular sulfate reduction rates were calculated. The rates were highest in the surface layer (0.14 fmol cell(-1) day(-1)), decreased by a factor of 3 within the first 2 cm, and were relatively constant in deeper layers.
Figures




References
-
- Canfield D E, Jørgensen B B, Fossing H, Glud R, Gundersen J, Ramsing N B, Thamdrup B, Hansen J W, Nielsen L P, Hall P O J. Pathways of organic carbon oxidation in three continental margin sediments. Mar Geol. 1993;113:27–40. - PubMed
-
- Coleman M L, Hedrick D B, Lovley D R, White D C, Pye K. Reduction of Fe(III) in sediments by sulphate-reducing bacteria. Nature. 1993;361:436–438.
-
- Dannenberg S, Kroder M, Dilling W, Cypionka H. Oxidation of H2, organic compounds and inorganic sulfur compounds coupled to reduction of O2 or nitrate by sulfate-reducing bacteria. Arch Microbiol. 1992;158:93–99.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

