A role for transcriptional repression of p21CIP1 by c-Myc in overcoming transforming growth factor beta -induced cell-cycle arrest
- PMID: 10920185
- PMCID: PMC16893
- DOI: 10.1073/pnas.150006697
A role for transcriptional repression of p21CIP1 by c-Myc in overcoming transforming growth factor beta -induced cell-cycle arrest
Abstract
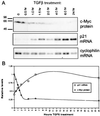
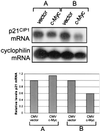
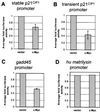
c-Myc plays a vital role in cell-cycle progression. Deregulated expression of c-Myc can overcome cell-cycle arrest in order to promote cellular proliferation. Transforming growth factor beta (TGFbeta) treatment of immortalized human keratinocyte cells inhibits cell-cycle progression and is characterized by down-regulation of c-Myc followed by up-regulation of p21(CIP1). A direct role of c-Myc in this pathway was demonstrated by the observation that ectopic expression of c-Myc overcame the cell-cycle block induced by TGFbeta treatment. The induction of p21(CIP1) transcription by TGFbeta was blocked in human keratinocyte cells stably expressing c-Myc. Furthermore, overexpression of c-Myc in NIH 3T3 cells repressed the basal levels of p21(CIP1) mRNA. Repression of p21(CIP1) transcription by c-Myc occurred at the promoter level in a region near the start site of transcriptional initiation and was independent of histone deacetylase activity. These data suggest that the down-regulation of c-Myc after TGFbeta signaling is important for subsequent regulation of p21(CIP1) and cell-cycle inhibition. Thus, repression of the cell-cycle inhibitory gene p21(CIP1) plays a role in c-Myc-dependent cell-cycle progression.
Figures



References
-
- Mateyak M K, Obaya A J, Adachi S, Sedivy J M. Cell Growth Diff. 1997;8:1039–1048. - PubMed
-
- Karn J, Watson J V, Lowe A D, Green S M, Vedeckis W. Oncogene. 1989;4:773–787. - PubMed
-
- Cole M D, McMahon S B. Oncogene. 1999;18:2916–2924. - PubMed
-
- Claassen G F, Hann S R. Oncogene. 1999;18:2925–2933. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

