How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene
- PMID: 10921891
- PMCID: PMC306607
- DOI: 10.1093/emboj/19.15.4111
How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene
Erratum in
- EMBO J 2000 Sep 1;19(17):4855
Abstract
Analysis of mRNA levels in cells that express or lack signal transducers and activators of transcription 1 (Stat1) reveals that Stat1 mediates the constitutive transcription of many genes. Expression of the low molecular mass polypeptide 2 (LMP2), which requires Stat1, has been studied in detail. The overlapping interferon consensus sequence 2/gamma-interferon-activated sequence (ICS-2/GAS) elements in the LMP2 promoter bind to interferon regulatory factor 1 (IRF1) and Stat1 and are occupied constitutively in vivo. The point mutant of Stat1, Y701F, which does not form dimers involving SH2-phosphotyrosine interactions, binds to the GAS element and supports LMP2 expression. Unphosphorylated Stat1 binds to IRF1 directly and we conclude that this complex uses the ICS-2/GAS element to mediate constitutive LMP2 transcription in vivo. The promoter of the IRF1 gene, which also contains a GAS site but not an adjacent ICS-2 site, is not activated by Stat1 Y701F. The promoters of other genes whose constitutive expression requires Stat1 may also utilize complexes of unphosphorylated Stat1 with IRF1 or other transcription factors.
Figures
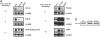
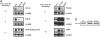







References
-
- Baden H.A., Sarma,S.P., Kapust,R.B., Byrd,R.A. and Waugh,D.S. (1998) The amino-terminal domain of human STAT4. Overproduction, purification, and biophysical characterization. J. Biol. Chem., 273, 17109–17114. - PubMed
-
- Chatterjee-Kishore M., Kishore,R., Hicklin,D.J., Marincola,F.M. and Ferrone,S. (1998) Different requirements for signal transducer and activator of transcription 1α and interferon regulatory factor 1 in the regulation of low molecular mass polypeptide 2 and transporter associated with antigen processing 1 gene expression. J. Biol. Chem., 273, 16177–16183. - PubMed
-
- Chatterjee-Kishore M., van den Akker,F. and Stark,G.R. (2000) Adenovirus E1A down-regulates LMP2 transcription by interfering with the binding of Stat1 to IRF1. J. Biol. Chem., 275, 20406–20411. - PubMed
-
- Chen X., Vinkemeier,U., Zhao,Y., Jeruzalmi,D., Darnell,J.E.,Jr and Kuriyan,J. (1998) Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell, 93, 827–839. - PubMed
-
- Darnell J.E. Jr, Kerr,I.M. and Stark,G.R. (1994) Jak–STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science, 264, 1415–1421. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

