Sip2p and its partner snf1p kinase affect aging in S. cerevisiae
- PMID: 10921902
- PMCID: PMC316829
Sip2p and its partner snf1p kinase affect aging in S. cerevisiae
Abstract
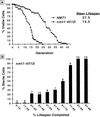
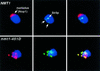
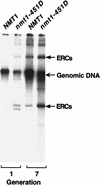
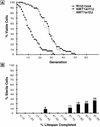
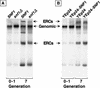
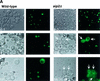
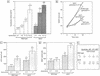
For a number of organisms, the ability to withstand periods of nutrient deprivation correlates directly with lifespan. However, the underlying molecular mechanisms are poorly understood. We show that deletion of the N-myristoylprotein, Sip2p, reduces resistance to nutrient deprivation and shortens lifespan in Saccharomyces cerevisiae. This reduced lifespan is due to accelerated aging, as defined by loss of silencing from telomeres and mating loci, nucleolar fragmentation, and accumulation of extrachromosomal rDNA. Genetic studies indicate that sip2Delta produces its effect on aging by increasing the activity of Snf1p, a serine/threonine kinase involved in regulating global cellular responses to glucose starvation. Biochemical analyses reveal that as yeast age, hexokinase activity increases as does cellular ATP and NAD(+) content. The change in glucose metabolism represents a new correlate of aging in yeast and occurs to a greater degree, and at earlier generational ages in sip2Delta cells. Sip2p and Snf1p provide new molecular links between the regulation of cellular energy utilization and aging.
Figures

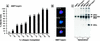


References
-
- Alepuz PM, Cunningham KW, Estruch F. Glucose repression affects ion homeostasis in yeast through the regulation of the stress-activated ENA1 gene. Mol Microbiol. 1997;26:91–98. - PubMed
-
- Arking R. Biology of Aging. 2nd Edition. Sunderland, MA: Sinaur Associates; 1998.
-
- Arking R, Buck S, Berrios A, Dwyer S, Baker GT., III Elevated paraquat resistance can be used as a bioassay for longevity in a genetically based long-lived strain of Drosophila. Dev Genet. 1991;12:362–370. - PubMed
-
- Ashrafi K, Farazi TA, Gordon JI. A role for Saccharomyces cerevisiae fatty acid activation protein 4 in regulating protein N-myristoylation during entry into stationary phase. J Biol Chem. 1998;273:25864–25874. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
