Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress
- PMID: 10921903
- PMCID: PMC316817
Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress
Abstract
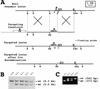
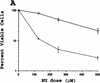
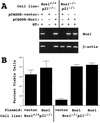
The eukaryotic cell cycle is overseen by regulatory mechanisms, termed checkpoints, that respond to DNA damage, mitotic spindle defects, and errors in the ordering of cell cycle events. The DNA replication and DNA damage cell cycle checkpoints of the fission yeast Schizosaccharomyces pombe require the hus1(+) (hydroxyurea sensitive) gene. To determine the role of the mouse homolog of hus1(+) in murine development and cell cycle checkpoint function, we produced a targeted disruption of mouse Hus1. Inactivation of Hus1 results in mid-gestational embryonic lethality due to widespread apoptosis and defective development of essential extra-embryonic tissues. DNA damage-inducible genes are up-regulated in Hus1-deficient embryos, and primary cells from Hus1-null embryos contain increased spontaneous chromosomal abnormalities, suggesting that loss of Hus1 leads to an accumulation of genome damage. Embryonic fibroblasts lacking Hus1 fail to proliferate in vitro, but inactivation of p21 allows for the continued growth of Hus1-deficient cells. Hus1(-/-)p21(-/-) cells display a unique profile of significantly heightened sensitivity to hydroxyurea, a DNA replication inhibitor, and ultraviolet light, but only slightly increased sensitivity to ionizing radiation. Taken together, these results indicate that mouse Hus1 functions in the maintenance of genomic stability and additionally identify an evolutionarily-conserved role for Hus1 in mediating cellular responses to genotoxins.
Figures
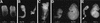






References
-
- Ahmed S, Hodgkin J. MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature. 2000;403:159–164. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
