Comparison and application of a novel genotyping method, semiautomated primer-specific and mispair extension analysis, and four other genotyping assays for detection of hepatitis C virus mixed-genotype infections
- PMID: 10921931
- PMCID: PMC87116
- DOI: 10.1128/JCM.38.8.2807-2813.2000
Comparison and application of a novel genotyping method, semiautomated primer-specific and mispair extension analysis, and four other genotyping assays for detection of hepatitis C virus mixed-genotype infections
Abstract
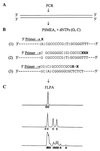
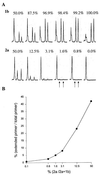
To date the true prevalence of hepatitis C virus (HCV) mixed-genotype infections has not been established mainly because currently available methods are not suitable for the detection of mixed genotypes in a viral population. A novel semiautomated genotyping method, primer-specific and mispair extension analysis (S-PSMEA), which is more reliable than other genotyping assays was developed for detection of HCV mixed-genotype infections. A genotype present at levels as low as 0.8% in a defined mix of HCV genotypes was detected, showing a 20-fold increase in sensitivity over that of direct DNA sequencing. A total of 434 HCV isolates were genotyped and analyzed for a comparative study of the accuracy between S-PSMEA and four current genotyping methods. The results showed that viruses in approximately 40% of the samples from this group determined to be infected with mixed genotypes by S-PSMEA were undetected by direct DNA sequencing due to its low sensitivity. Type-specific PCR, line probe assay, and restriction fragment length polymorphism analysis performed poorly, being able to identify only 38.5, 16.1, and 15.4% of mixed-genotype infections, respectively, that were detected by direct DNA sequencing. The prevalence of mixed-genotype infections detected by S-PSMEA was 7.9% (12 of 152 donors) among HCV-infected blood donors, 14.3% (15 of 105) among patients with chronic hepatitis C, and 17.1% (6 of 36) among thalassemia patients who had received multiple transfusions. The data lead us to conclude that HCV mixed-genotype infections are more common than previously estimated and that S-PSMEA may be the method of choice when detection of genotypes present at low levels in mixed-genotype infections is required due to its higher level of sensitivity.
Figures

Similar articles
-
Evaluation of three different hepatitis C virus typing methods for detection of mixed-genotype infections.J Dig Dis. 2011 Jun;12(3):199-203. doi: 10.1111/j.1751-2980.2011.00496.x. J Dig Dis. 2011. PMID: 21615874
-
Primer specific and mispair extension analysis (PSMEA) as a simple approach to fast genotyping.Nucleic Acids Res. 1998 Nov 1;26(21):5013-5. doi: 10.1093/nar/26.21.5013. Nucleic Acids Res. 1998. PMID: 9776770 Free PMC article.
-
Genotyping of hepatitis C virus isolates by a modified polymerase chain reaction assay using type specific primers: epidemiological applications.J Med Virol. 1994 Nov;44(3):272-9. doi: 10.1002/jmv.1890440311. J Med Virol. 1994. PMID: 7531757
-
Genotypes of hepatitis C virus isolates from different parts of the world.Arch Virol Suppl. 1996;11:185-93. doi: 10.1007/978-3-7091-7482-1_16. Arch Virol Suppl. 1996. PMID: 8800799 Review.
-
Genotyping & diagnostic methods for hepatitis C virus: A need of low-resource countries.Indian J Med Res. 2018 May;147(5):445-455. doi: 10.4103/ijmr.IJMR_1850_16. Indian J Med Res. 2018. PMID: 30082568 Free PMC article. Review.
Cited by
-
Effect of oligonucleotide primers in determining viral variability within hosts.Virol J. 2004 Dec 9;1:13. doi: 10.1186/1743-422X-1-13. Virol J. 2004. PMID: 15588294 Free PMC article.
-
Surveillance of viral contamination of invasive medical instruments in dentistry.J Zhejiang Univ Sci B. 2006 Sep;7(9):745-8. doi: 10.1631/jzus.2006.B0745. J Zhejiang Univ Sci B. 2006. PMID: 16909477 Free PMC article.
-
A shift in the hepatitis C virus genotype dominance in blood donor samples from Thailand.Mol Biol Rep. 2011 Oct;38(7):4287-90. doi: 10.1007/s11033-010-0552-x. Epub 2010 Nov 27. Mol Biol Rep. 2011. PMID: 21113670
-
A reliable multiplex genotyping assay for HCV using a suspension bead array.Microb Biotechnol. 2015 Jan;8(1):93-102. doi: 10.1111/1751-7915.12140. Epub 2014 Jul 10. Microb Biotechnol. 2015. PMID: 25042084 Free PMC article.
-
Molecular epidemiology of hepatitis C among drug users in Flanders, Belgium: association of genotype with clinical parameters and with sex- and drug-related risk behaviours.Eur J Clin Microbiol Infect Dis. 2005 Aug;24(8):514-22. doi: 10.1007/s10096-005-1376-9. Eur J Clin Microbiol Infect Dis. 2005. PMID: 16133411
References
-
- Alter M J. Epidemiology of hepatitis C. Hepatology. 1997;26:62S–65S. - PubMed
-
- Alter M J, Kruszon-Moran D, Nainan O V, McQuillan G M, Gao F, Moyer L A, Kaslow R A, Margolis H S. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N Engl J Med. 1999;341:556–562. - PubMed
-
- Bukh J, Miller R H, Purcell R H. Genetic heterogeneity of the hepatitis C virus: quasispecies and genotypes. Semin Liver Dis. 1995;15:41–63. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

