Genetic diversity, distribution, and serological features of hantavirus infection in five countries in South America
- PMID: 10921972
- PMCID: PMC87178
- DOI: 10.1128/JCM.38.8.3029-3035.2000
Genetic diversity, distribution, and serological features of hantavirus infection in five countries in South America
Abstract
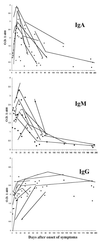
Since 1995 when the first case of hantavirus pulmonary syndrome (HPS) was reported in Patagonia, there have been more than 400 cases of HPS reported in five countries in South America. The first case of HPS was associated with Andes (AND) virus. In this study, we report on the genetic diversity, geographical distribution, and serological features of hantavirus infection in six countries in South America based on 87 HPS cases from Argentina, Bolivia, Chile, Paraguay, and Uruguay. An early immunoglobulin M (IgM), IgA, and IgG humoral response was observed in almost all HPS cases. The IgM response appears to peak 1 or 2 days after the onset of symptoms. Peak IgG antibody titers occur mostly after the first week. Low IgG titers or the absence of IgG was associated with higher mortality rates. The IgA response peaks around day 15 and then rapidly decreases. The results of phylogenetic analysis based on partial M-fragment G1- and G2-encoding sequences showed that HPS cases from the five countries were infected with viruses related to AND or Laguna Negra (LN) virus. Within AND virus-infected persons, at least five major genetic lineages were found; one lineage was detected in Uruguayan and Argentinean cases from both sides of the Rio de la Plata river. Two Paraguayan patients were infected with a virus different from LN virus. According to the results of phylogenetic analyses, this virus probably belongs to a distinct lineage related more closely to the AND virus than to the LN virus, suggesting that there is probably an Oligoryzomys-borne viral variant circulating in Paraguay. These studies may contribute to a better understanding of hantavirus human infection in South America.
Figures


References
-
- Bharadwaj M, Botten J, Torrez-Martinez N, Hjelle B. Rio Mamore virus: genetic characterization of a newly recognized hantavirus of the pygmy rice rat, Oligoryzomys microtis, from Bolivia. Am J Trop Med Hyg. 1997;57:368–374. - PubMed
-
- Calderon G, Pini N C, Bolpe J, Levis S, Mills J, Segura E L, Guthmann N, Cantoni G, Becker J, Fonollat A, Ripoll C, Bortman M, Benedetti R, Sabattini M, Enria D A. Hantavirus reservoir hosts associated with peridomestic habitants in Argentina. Emerg. Infect. Dis. Vol. 5. 1999. http://www.cdc.gov/ncidod/EID/vol5no6/calderon.htm [Online.] http://www.cdc.gov/ncidod/EID/vol5no6/calderon.htm. . - PMC - PubMed
-
- Chiappero M B, Calderon G E, Gardenal C N. Oligoryzomys flavescens (Rodentia, Muridae): gene flow among populations from central-eastern Argentina. Genetica. 1997;101:105–113. - PubMed
-
- Duchin J S, Koster F T, Peters C J, Simpson G L, Tempest B, Zaki S R, Ksiazek T G, Rollin P E, Nichol S, Umland E T, Moolenaar R L, Reef S E, Nolte K B, Gallher M M, Butler J C, Breiman R F The Hantavirus Study Group. Hantavirus pulmonary syndrome: a clinical description of 17 patients with a newly recognized disease. N Engl J Med. 1994;330:949–955. - PubMed
-
- Elliott R M, Schmaljohn C S, Collet M S. Bunyavirus genome structure and gene expression. Curr Top Microbiol Immunol. 1991;169:91–141. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

