Genetic complexity of pathogen perception by plants: the example of Rcr3, a tomato gene required specifically by Cf-2
- PMID: 10922039
- PMCID: PMC34016
- DOI: 10.1073/pnas.97.16.8807
Genetic complexity of pathogen perception by plants: the example of Rcr3, a tomato gene required specifically by Cf-2
Abstract
Genetic analysis of plant-pathogen interactions has demonstrated that resistance to infection is often determined by the interaction of dominant plant resistance (R) genes and dominant pathogen-encoded avirulence (Avr) genes. It was postulated that R genes encode receptors for Avr determinants. A large number of R genes and their cognate Avr genes have now been analyzed at the molecular level. R gene loci are extremely polymorphic, particularly in sequences encoding amino acids of the leucine-rich repeat motif. A major challenge is to determine how Avr perception by R proteins triggers the plant defense response. Mutational analysis has identified several genes required for the function of specific R proteins. Here we report the identification of Rcr3, a tomato gene required specifically for Cf-2-mediated resistance. We propose that Avr products interact with host proteins to promote disease, and that R proteins "guard" these host components and initiate Avr-dependent plant defense responses.
Figures


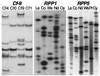
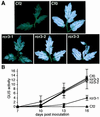

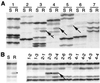
References
-
- Flor H H. Annu Rev Phytopathol. 1971;9:275–296.
-
- Galan E J, Collmer A. Science. 1999;284:1322–1328. - PubMed
-
- Kearney B, Staskawicz B J. Nature (London) 1990;346:385–386. - PubMed
-
- Ritter C, Dangl J L. Mol Plant-Microbe Interact. 1995;8:444–453. - PubMed
-
- Erickson F L, Holzberg S, Calderon-Urrea A, Handley V, Axtell M, Corr C, Baker B. Plant J. 1999;18:67–75. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

