Hydrolytic editing by a class II aminoacyl-tRNA synthetase
- PMID: 10922054
- PMCID: PMC16796
- DOI: 10.1073/pnas.97.16.8916
Hydrolytic editing by a class II aminoacyl-tRNA synthetase
Abstract
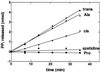
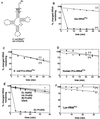
Editing reactions catalyzed by aminoacyl-tRNA synthetases are critical for accurate translation of the genetic code. To date, this activity, whereby misactivated amino acids are hydrolyzed either before or after transfer to noncognate tRNAs, has been characterized extensively only in the case of class I synthetases. Class II synthetases have an active-site architecture that is completely distinct from that of class I. Thus, findings on editing by class I synthetases may not be applicable generally to class II enzymes. Class II Escherichia coli proline-tRNA synthetase is shown here to misactivate alanine and to hydrolyze the noncognate amino acid before transfer to tRNA(Pro). This enzyme also is capable of rapidly deacylating a mischarged Ala-tRNA(Pro) variant. A single cysteine residue (C443) that is located within the class II-specific motif 3 consensus sequence was shown previously to be dispensable for proline-tRNA synthetase aminoacylation activity. We show here that C443 is critical for the hydrolytic editing of Ala-tRNA(Pro) by this class II synthetase.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

