Energetics of the HIV gp120-CD4 binding reaction
- PMID: 10922058
- PMCID: PMC16815
- DOI: 10.1073/pnas.97.16.9026
Energetics of the HIV gp120-CD4 binding reaction
Abstract
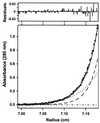
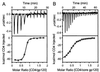
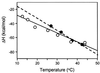
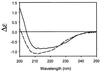
HIV infection is initiated by the selective interaction between the cellular receptor CD4 and gp120, the external envelope glycoprotein of the virus. We used analytical ultracentrifugation, titration calorimetry, and surface plasmon resonance biosensor analysis to characterize the assembly state, thermodynamics, and kinetics of the CD4-gp120 interaction. The binding thermodynamics were of unexpected magnitude; changes in enthalpy, entropy, and heat capacity greatly exceeded those described for typical protein-protein interactions. These unusual thermodynamic properties were observed with both intact gp120 and a deglycosylated and truncated form of gp120 protein that lacked hypervariable loops V1, V2, and V3 and segments of its N and C termini. Together with previous crystallographic studies, the large changes in heat capacity and entropy reveal that extensive structural rearrangements occur within the core of gp120 upon CD4 binding. CD spectral studies and slow kinetics of binding support this conclusion. These results indicate considerable conformational flexibility within gp120, which may relate to viral mechanisms for triggering infection and disguising conserved receptor-binding sites from the immune system.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

