Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes
- PMID: 10931856
- PMCID: PMC2175199
- DOI: 10.1083/jcb.150.3.405
Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes
Abstract
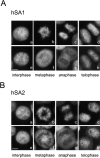
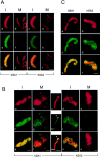
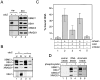
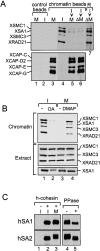
A multisubunit protein complex, termed cohesin, plays an essential role in sister chromatid cohesion in yeast and in Xenopus laevis cell-free extracts. We report here that two distinct cohesin complexes exist in Xenopus egg extracts. A 14S complex (x-cohesin(SA1)) contains XSMC1, XSMC3, XRAD21, and a newly identified subunit, XSA1. In a second 12.5S complex (x-cohesin(SA2)), XSMC1, XSMC3, and XRAD21 associate with a different subunit, XSA2. Both XSA1 and XSA2 belong to the SA family of mammalian proteins and exhibit similarity to Scc3p, a recently identified component of yeast cohesin. In Xenopus egg extracts, x-cohesin(SA1) is predominant, whereas x-cohesin(SA2) constitutes only a very minor population. Human cells have a similar pair of cohesin complexes, but the SA2-type is the dominant form in somatic tissue culture cells. Immunolocalization experiments suggest that chromatin association of cohesin(SA1) and cohesin(SA2) may be differentially regulated. Dissociation of x-cohesin(SA1) from chromatin correlates with phosphorylation of XSA1 in the cell-free extracts. Purified cdc2-cyclin B can phosphorylate XSA1 in vitro and reduce the ability of x-cohesin(SA1) to bind to DNA or chromatin. These results shed light on the mechanism by which sister chromatid cohesion is partially dissolved in early mitosis, far before the onset of anaphase, in vertebrate cells.
Figures




References
-
- Bell S.P., Kobayashi R., Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993;262:1844–1849. - PubMed
-
- Biggins S., Murray A. Sister chromatid cohesion in mitosis. Curr. Opin. Cell Biol. 1999;9:230–236. - PubMed
-
- Birkenbihl R.P., Subramani S. The rad21 gene product of Schizosaccharomyces pombe is a nuclear, cell cycle-regulated phosphoprotein. J. Biol. Chem. 1995;270:7703–7711. - PubMed
-
- Blat Y., Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. - PubMed
-
- Carramolino L., Lee B.C., Zaballos A., Peled A., Barthelemy I., Shav-Tal Y., Prieto I., Carmi P., Gothelf Y., González de Buitrago G. SA-1, a nuclear protein encoded by one member of a novel gene familymolecular cloning and detection in hemopoietic organs. Gene. 1997;195:151–159. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

