TARP: a nuclear protein expressed in prostate and breast cancer cells derived from an alternate reading frame of the T cell receptor gamma chain locus
- PMID: 10931945
- PMCID: PMC16882
- DOI: 10.1073/pnas.160270597
TARP: a nuclear protein expressed in prostate and breast cancer cells derived from an alternate reading frame of the T cell receptor gamma chain locus
Abstract
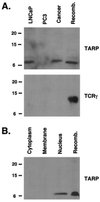
Previously, we identified the expression of a prostate-specific form of T cell receptor gamma chain (TCRgamma) mRNA in the human prostate and demonstrated that it originates from epithelial cells and not from infiltrating T lymphocytes. Here, we show that this prostate-specific transcript is also expressed in three breast cancer cell lines and breast cancer tissues. Analysis of the cDNA sequence predicts that this transcript can encode two protein products of 7 and 13 kDa, and in vitro translation experiments showed that both proteins were made. The longer ORF encodes a 13-kDa truncated version of TCRgamma, whereas the shorter alternative reading frame encodes a 7-kDa protein with five leucine residues in heptad repeats followed by a basic region. Studies with specific antibodies against each protein product revealed that both prostate and breast cancer cells contain only the 7-kDa protein, which is located in the nucleus. We have named this protein TCRgamma alternate reading frame protein (TARP). These results demonstrate that an alternative protein product is encoded by the TCRgamma locus in cells other than T lymphocytes.
Figures






References
-
- Hara T, Harada N, Mitsui H, Miura T, Ishizaka T, Miyajima A. Blood. 1994;84:189–199. - PubMed
-
- Liang P, Pardee A B. Science. 1992;257:967–971. - PubMed
-
- Velculescu V E, Zhang L, Vogelstein B, Kinzler K W. Science. 1995;276:1268–1272. - PubMed
-
- Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

