Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana
- PMID: 10931949
- PMCID: PMC16942
- DOI: 10.1073/pnas.160276997
Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana
Abstract
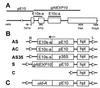
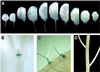
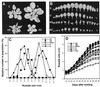
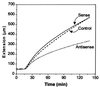
Expansins are cell-wall-loosening proteins that induce stress relaxation and extension of plant cell walls. To evaluate their hypothesized role in cell growth, we genetically manipulated expansin gene expression in Arabidopsis thaliana and assessed the consequent changes in growth and cell-wall properties. Various combinations of promoters were used to drive antisense and sense sequences of AtEXP10, which is maximally expressed in the growing leaf and at the base of the pedicel. Compared with controls, antisense lines had smaller rosettes because of shorter petioles and leaf blades and often acquired a twisted leaf morphology. Petiole cells from antisense plants were smaller than controls and their cell walls were significantly less extensible in vitro. Sense plants had slightly longer petioles, larger leaf blades, and larger cells than controls. Abscission at the base of the pedicel, where AtEXP10 is endogenously expressed, was enhanced in sense plants but reduced in antisense lines. These results support the concept that expansins function endogenously as cell-wall-loosening agents and indicate that expansins have versatile developmental roles that include control of organ size, morphology, and abscission.
Figures



References
-
- Cosgrove D J. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:391–417. - PubMed
-
- Carpita N C, Gibeaut D M. Plant J. 1993;3:1–30. - PubMed
-
- Gonzalez-Carranza Z H, Roberts J A, Lozoya-Gloria E. Trends Plant Sci. 1998;3:10–14.
-
- Rose J K C, Bennett A B. Trends Plant Sci. 1999;4:176–183. - PubMed
-
- Brummell D A, Hall B D, Bennett A B. Plant Mol Biol. 1999;40:615–622. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

