HSV-1-based vectors for gene therapy of neurological diseases and brain tumors: part I. HSV-1 structure, replication and pathogenesis
- PMID: 10933054
- PMCID: PMC1508113
- DOI: 10.1038/sj.neo.7900055
HSV-1-based vectors for gene therapy of neurological diseases and brain tumors: part I. HSV-1 structure, replication and pathogenesis
Abstract
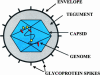
The design of effective gene therapy strategies for brain tumors and other neurological disorders relies on the understanding of genetic and pathophysiological alterations associated with the disease, on the biological characteristics of the target tissue, and on the development of safe vectors and expression systems to achieve efficient, targeted and regulated, therapeutic gene expression. The herpes simplex virus type 1 (HSV-1) virion is one of the most efficient of all current gene transfer vehicles with regard to nuclear gene delivery in central nervous system-derived cells including brain tumors. HSV-1-related research over the past decades has provided excellent insight into the structure and function of this virus, which, in turn, facilitated the design of innovative vector systems. Here, we review aspects of HSV-1 structure, replication and pathogenesis, which are relevant for the engineering of HSV-1-based vectors.
Figures


Similar articles
-
HSV-1-based vectors for gene therapy of neurological diseases and brain tumors: part II. Vector systems and applications.Neoplasia. 1999 Nov;1(5):402-16. doi: 10.1038/sj.neo.7900056. Neoplasia. 1999. PMID: 10933055 Free PMC article. Review.
-
HSV trafficking and development of gene therapy vectors with applications in the nervous system.Gene Ther. 2005 Jun;12(11):891-901. doi: 10.1038/sj.gt.3302545. Gene Ther. 2005. PMID: 15908995 Review.
-
Gene transfer to neurons using herpes simplex virus-based vectors.Annu Rev Neurosci. 1996;19:265-87. doi: 10.1146/annurev.ne.19.030196.001405. Annu Rev Neurosci. 1996. PMID: 8833444 Review.
-
Replication-defective genomic herpes simplex vectors: design and production.Curr Opin Biotechnol. 2002 Oct;13(5):424-8. doi: 10.1016/s0958-1669(02)00359-2. Curr Opin Biotechnol. 2002. PMID: 12459332 Review.
-
Overview of gene delivery into cells using HSV-1-based vectors.Curr Protoc Neurosci. 2012 Oct;Chapter 4:Unit 4.12. doi: 10.1002/0471142301.ns0412s61. Curr Protoc Neurosci. 2012. PMID: 23093351
Cited by
-
Antiviral Bioactive Compounds of Mushrooms and Their Antiviral Mechanisms: A Review.Viruses. 2021 Feb 23;13(2):350. doi: 10.3390/v13020350. Viruses. 2021. PMID: 33672228 Free PMC article. Review.
-
PET-based molecular imaging in neuroscience.Eur J Nucl Med Mol Imaging. 2003 Jul;30(7):1051-65. doi: 10.1007/s00259-003-1202-5. Epub 2003 May 23. Eur J Nucl Med Mol Imaging. 2003. PMID: 12764552 Review.
-
Oncolytic Virus Encoding a Master Pro-Inflammatory Cytokine Interleukin 12 in Cancer Immunotherapy.Cells. 2020 Feb 10;9(2):400. doi: 10.3390/cells9020400. Cells. 2020. PMID: 32050597 Free PMC article. Review.
-
Glioblastoma multiforme: State of the art and future therapeutics.Surg Neurol Int. 2014 May 8;5:64. doi: 10.4103/2152-7806.132138. eCollection 2014. Surg Neurol Int. 2014. PMID: 24991467 Free PMC article. Review.
-
Detection of spontaneous schwannomas by MRI in a transgenic murine model of neurofibromatosis type 2.Neoplasia. 2002 Nov-Dec;4(6):501-9. doi: 10.1038/sj.neo.7900265. Neoplasia. 2002. PMID: 12407444 Free PMC article.
References
-
- Young AB. Neurological Disorders: An Overwiew. In: Chiocca EA, Breakefield XO, editors. Gene Therapy for Neurological Disorders and Brain Tumors. Humana Press; 1998. pp. 341–343.
-
- Isacson O, Breakefield XO. Benefits and risks of hosting animal cells in the human brain. Nat Med. 1997;3:964–969. - PubMed
-
- Roizman B, Sears AE. Herpes Simplex Viruses and Their Replication. In: Fields BN, Knipe DM, Howley PM, et al., editors. Fields Virology. Philadelphia, PA: Lippincott; 1996. pp. 2231–2295.
-
- McGeoch DJ, Dolan A, Donald S, Rixon FJ. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J Mol Biol. 1985;181:1–13. - PubMed
-
- McGeoch DJ, Dalrymple MA, Davison AJ, Dolan A, Frame MC, McNab D, Perry LJ, Scott JE, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
