Existence of distinct GB virus C/hepatitis G virus variants with different tropism
- PMID: 10933701
- PMCID: PMC112324
- DOI: 10.1128/jvi.74.17.7936-7942.2000
Existence of distinct GB virus C/hepatitis G virus variants with different tropism
Abstract
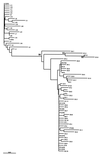
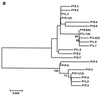
To study the existence of GB virus C/hepatitis G virus (GBV-C/HGV) variants with different tropism, we have analyzed the heterogeneity and quasispecies composition of GBV-C/HGV isolated from in vitro-infected peripheral blood mononuclear cells (PBMC) and from sera, livers, and PBMC from two chronically infected patients. For this purpose, the GBV-C/HGV 5' noncoding region (5'NCR) was amplified by reverse transcription-PCR and the amplified products were cloned and sequenced. These analyses showed that the master 5'NCR sequences isolated from the in vitro-infected PBMC and from the PBMC isolated from the patient whose serum was used as the inoculum were identical but different from that of the inoculum. Furthermore, phylogenetic analysis revealed that all PBMC sequences grouped together into a branch which was separate from those of the inoculum. For one of the two chronically infected patients, all the sequences from the PBMC and one from the liver clustered into a single branch while the sequences from the serum and all the other liver sequences grouped together in the other branch. For the other patient, the sequences from the serum and PBMC and three sequences from the liver grouped together into one branch, while the remaining five sequences from the liver were separated in a different cluster. In conclusion, our results support the existence of different GBV-C/HGV variants with different tissue tropism.
Figures



References
-
- Aikawa T, Sugay Y, Okamoto H. Hepatitis G infection in drug abusers with chronic hepatitis C. N Engl J Med. 1996;334:195. - PubMed
-
- Alter H J, Nakatsuyi Y, Melpolder J, Wages J, Wesley R, Shih J W K, Kim J P. The incidence of transfusion-associated hepatitis G virus infection and its relation to liver disease. N Engl J Med. 1997;336:747–754. - PubMed
-
- Alter M J, Gallagher M, Morris T T, Moyer L A, Meeks E L, Krawczynski K, Kim J P, Margolis H S. Acute non-A-E hepatitis in the United States and the role of hepatitis G virus infection. N Engl J Med. 1997;336:741–746. - PubMed
-
- Berenguer M, Terrault N A, Piatak M, Yun A, Kim J P, Lau J Y N, Lake J R, Roberts J R, Ascher N, Ferrel L, Wright T. Hepatitis G virus infection in patients with hepatitis C virus infection undergoing liver transplantation. Gastroenterology. 1996;111:1569–1575. - PubMed
-
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

