A second leukotriene B(4) receptor, BLT2. A new therapeutic target in inflammation and immunological disorders
- PMID: 10934230
- PMCID: PMC2193217
- DOI: 10.1084/jem.192.3.421
A second leukotriene B(4) receptor, BLT2. A new therapeutic target in inflammation and immunological disorders
Abstract
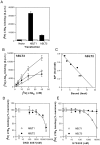
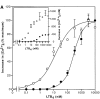
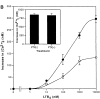
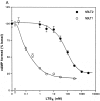
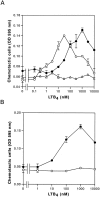
Leukotriene B(4) (LTB(4)) is a potent chemoattractant and activator of both granulocytes and macrophages. The actions of LTB(4) appear to be mediated by a specific G protein-coupled receptor (GPCR) BLT1, originally termed BLT (Yokomizo, T., T. Izumi, K. Chang, Y. Takuwa, and T. Shimizu. 1997. Nature. 387:620-624). Here, we report the molecular cloning of a novel GPCR for LTB(4), designated BLT2, which binds LTB(4) with a Kd value of 23 nM compared with 1.1 nM for BLT1, but still efficiently transduces intracellular signaling. BLT2 is highly homologous to BLT1, with an amino acid identity of 45.2%, and its open reading frame is located in the promoter region of the BLT1 gene. BLT2 is expressed ubiquitously, in contrast to BLT1, which is expressed predominantly in leukocytes. Chinese hamster ovary cells expressing BLT2 exhibit LTB(4)-induced chemotaxis, calcium mobilization, and pertussis toxin-insensitive inhibition of adenylyl cyclase. Several BLT1 antagonists, including U 75302, failed to inhibit LTB(4) binding to BLT2. Thus, BLT2 is a pharmacologically distinct receptor for LTB(4), and may mediate cellular functions in tissues other than leukocytes. BLT2 provides a novel target for antiinflammatory therapy and promises to expand our knowledge of LTB(4) function. The location of the gene suggests shared transcriptional regulation of these two receptors.
Figures
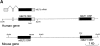




Comment in
-
The scent of a phagocyte: Advances on leukotriene b(4) receptors.J Exp Med. 2000 Aug 7;192(3):F5-8. doi: 10.1084/jem.192.3.f5. J Exp Med. 2000. PMID: 10934235 Free PMC article. Review. No abstract available.
References
-
- Samuelsson B., Dahlén S.E., Lindgren J.Å., Rouzer C.A., Serhan C.N. Leukotrienes and lipoxinsstructures, biosynthesis, and biological effects. Science. 1987;237:1171–1176. - PubMed
-
- Shimizu T., Wolfe L.S. Arachidonic acid cascade and signal transduction. J. Neurochem. 1990;55:1–15. - PubMed
-
- Serhan C.N., Haeggstrom J.Z., Leslie C.C. Lipid mediator networks in cell signalingupdate and impact of cytokines. FASEB J. 1996;10:1147–1158. - PubMed
-
- Iversen L., Kragballe K., Ziboh V.A. Significance of leukotriene-A4 hydrolase in the pathogenesis of psoriasis. Skin. Pharmacol. 1997;10:169–177. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

