BLTR mediates leukotriene B(4)-induced chemotaxis and adhesion and plays a dominant role in eosinophil accumulation in a murine model of peritonitis
- PMID: 10934232
- PMCID: PMC2193216
- DOI: 10.1084/jem.192.3.439
BLTR mediates leukotriene B(4)-induced chemotaxis and adhesion and plays a dominant role in eosinophil accumulation in a murine model of peritonitis
Abstract
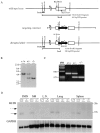
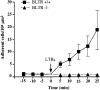
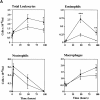
Leukotriene B(4) (LTB(4)) is a potent chemoattractant active on multiple leukocytes, including neutrophils, macrophages, and eosinophils, and is implicated in the pathogenesis of a variety of inflammatory processes. A seven transmembrane-spanning, G protein-coupled receptor, called BLTR (LTB(4) receptor), has recently been identified as an LTB(4) receptor. To determine if BLTR is the sole receptor mediating LTB(4)-induced leukocyte activation and to determine the role of LTB(4) and BLTR in regulating leukocyte function in inflammation in vivo, we generated a BLTR-deficient mouse by targeted gene disruption. This mouse reveals that BLTR alone is responsible for LTB(4)-mediated leukocyte calcium flux, chemotaxis, and firm adhesion to endothelium in vivo. Furthermore, despite the apparent functional redundancy with other chemoattractant-receptor pairs in vitro, LTB(4) and BLTR play an important role in the recruitment and/or retention of leukocytes, particularly eosinophils, to the inflamed peritoneum in vivo. These studies demonstrate that BLTR is the key receptor that mediates LTB(4)-induced leukocyte activation and establishes a model to decipher the functional roles of BLTR and LTB(4) in vivo.
Figures

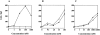
Comment in
-
The scent of a phagocyte: Advances on leukotriene b(4) receptors.J Exp Med. 2000 Aug 7;192(3):F5-8. doi: 10.1084/jem.192.3.f5. J Exp Med. 2000. PMID: 10934235 Free PMC article. Review. No abstract available.
References
-
- Samuelsson B., Dahlen S.E., Lindgren J.A., Rouzer C.A., Serhan C.N. Leukotrienes and lipoxinsstructures, biosynthesis, and biological effects. Science. 1987;237:1171–1176. - PubMed
-
- Lewis R.A., Austen K.F., Soberman R.J. Leukotrienes and other products of the 5-lipoxygenase pathway. Biochemistry and relation to pathobiology in human diseases. N. Engl. J. Med. 1990;323:645–655. - PubMed
-
- Ford-Hutchinson A.W., Bray M.A., Doig M.V., Shipley M.E., Smith M. Leukotriene B4, a potent chemokinetic and aggregating substance released from polymorphonuclear leukocytes. Nature. 1980;286:264–265. - PubMed
-
- Ng C.F., Sun F.F., Taylor B.M., Wolin M.S., Wong P.Y.-K. Functional properties of guinea pig eosinophil leukotriene B4 receptor. J. Immunol. 1991;147:3096–3103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

