Targeted disruption of the mPer3 gene: subtle effects on circadian clock function
- PMID: 10938103
- PMCID: PMC86101
- DOI: 10.1128/MCB.20.17.6269-6275.2000
Targeted disruption of the mPer3 gene: subtle effects on circadian clock function
Abstract
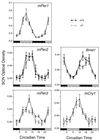
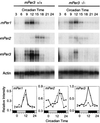
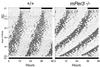
Neurons in the mammalian suprachiasmatic nucleus (SCN) contain a cell-autonomous circadian clock that is based on a transcriptional-translational feedback loop. The basic helix-loop-helix-PAS proteins CLOCK and BMAL1 are positive regulators and drive the expression of the negative regulators CRY1 and CRY2, as well as PER1, PER2, and PER3. To assess the role of mouse PER3 (mPER3) in the circadian timing system, we generated mice with a targeted disruption of the mPer3 gene. Western blot analysis confirmed the absence of mPER3-immunoreactive proteins in mice homozygous for the targeted allele. mPer1, mPer2, mCry1, and Bmal1 RNA rhythms in the SCN did not differ between mPER3-deficient and wild-type mice. Rhythmic expression of mPer1 and mPer2 RNAs in skeletal muscle also did not differ between mPER3-deficient and wild-type mice. mPer3 transcripts were rhythmically expressed in the SCN and skeletal muscle of mice homozygous for the targeted allele, but the level of expression of the mutant transcript was lower than that in wild-type controls. Locomotor activity rhythms in mPER3-deficient mice were grossly normal, but the circadian cycle length was significantly (0.5 h) shorter than that in controls. The results demonstrate that mPer3 is not necessary for circadian rhythms in mice.
Figures



References
-
- Akiyama M, Kouzu Y, Takahashi S, Wakamatsu H, Moriya T, Maetani M, Watanabe S, Tei H, Sakaki Y, Shibata S. Inhibition of light- or glutamate-induced mPer1 expression represses the phase shifts into the mouse circadian locomotor and suprachiasmatic firing rhythms. J Neurosci. 1999;19:1115–1121. - PMC - PubMed
-
- Albrecht U, Sun Z S, Eichele G, Lee C C. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. - PubMed
-
- Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. - PubMed
-
- Dunlap J C. Molecular bases for circadian clocks. Cell. 1999;96:271–290. - PubMed
-
- Field M D, Maywood E S, O'Brien J A, Weaver D R, Reppert S M, Hastings M H. Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron. 2000;25:437–447. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
