Functional studies on the candidate ATPase domains of Saccharomyces cerevisiae MutLalpha
- PMID: 10938116
- PMCID: PMC86114
- DOI: 10.1128/MCB.20.17.6390-6398.2000
Functional studies on the candidate ATPase domains of Saccharomyces cerevisiae MutLalpha
Abstract
Saccharomyces cerevisiae MutL homologues Mlh1p and Pms1p form a heterodimer, termed MutLalpha, that is required for DNA mismatch repair after mismatch binding by MutS homologues. Recent sequence and structural studies have placed the NH(2) termini of MutL homologues in a new family of ATPases. To address the functional significance of this putative ATPase activity in MutLalpha, we mutated conserved motifs for ATP hydrolysis and ATP binding in both Mlh1p and Pms1p and found that these changes disrupted DNA mismatch repair in vivo. Limited proteolysis with purified recombinant MutLalpha demonstrated that the NH(2) terminus of MutLalpha undergoes conformational changes in the presence of ATP and nonhydrolyzable ATP analogs. Furthermore, two-hybrid analysis suggested that these ATP-binding-induced conformational changes promote an interaction between the NH(2) termini of Mlh1p and Pms1p. Surprisingly, analysis of specific mutants suggested differential requirements for the ATPase motifs of Mlh1p and Pms1p during DNA mismatch repair. Taken together, these results suggest that MutLalpha undergoes ATP-dependent conformational changes that may serve to coordinate downstream events during yeast DNA mismatch repair.
Figures
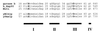
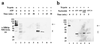


References
-
- Alani E, Chi N-W, Kolodner R. The Saccharomyces cerevisiae Msh2 protein specifically binds to duplex oligonucleotides containing mismatched DNA base pairs and insertions. Genes Dev. 1995;9:234–247. - PubMed
-
- Ali J A, Jackson A P, Howells A J, Maxwell A. The 43-kilodalton N-terminal fragment of the DNA gyrase b protein hydrolyzes ATP and binds coumarin drugs. Biochemistry. 1993;32:2717–2724. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
