A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition
- PMID: 10938129
- PMCID: PMC86127
- DOI: 10.1128/MCB.20.17.6537-6549.2000
A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition
Abstract
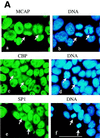
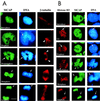
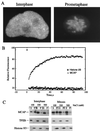
We describe a novel nuclear factor called mitotic chromosome-associated protein (MCAP), which belongs to the poorly understood BET subgroup of the bromodomain superfamily. Expression of the 200-kDa MCAP was linked to cell division, as it was induced by growth stimulation and repressed by growth inhibition. The most notable feature of MCAP was its association with chromosomes during mitosis, observed at a time when the majority of nuclear regulatory factors were released into the cytoplasm, coinciding with global cessation of transcription. Indicative of its predominant interaction with euchromatin, MCAP localized on mitotic chromosomes with exquisite specificity: (i) MCAP-chromosome association became evident subsequent to the initiation of histone H3 phosphorylation and early chromosomal condensation; and (ii) MCAP was absent from centromeres, the sites of heterochromatin. Supporting a role for MCAP in G(2)/M transition, microinjection of anti-MCAP antibody into HeLa cell nuclei completely inhibited the entry into mitosis, without abrogating the ongoing DNA replication. These results suggest that MCAP plays a role in a process governing chromosomal dynamics during mitosis.
Figures








References
-
- Beck S, Hanson I, Kelly A, Pappin D J, Trowsdale J. A homologue of the Drosophila female sterile homeotic (fsh) gene in the class II region of the human MHC. DNA Seq. 1992;2:203–210. - PubMed
-
- Coligan J, Kruisbeek A, Shevach E, Margulies D, editors. Current protocols in immunology. New York, N.Y: John Wiley & Sons; 1997.
-
- Coverley D, Laskey R A. Regulation of eukaryotic DNA replication. Annu Rev Biochem. 1994;63:745–776. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
