Limited correlation between expansin gene expression and elongation growth rate
- PMID: 10938357
- PMCID: PMC59097
- DOI: 10.1104/pp.123.4.1399
Limited correlation between expansin gene expression and elongation growth rate
Abstract
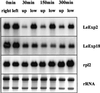
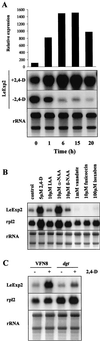
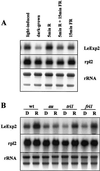
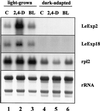
The aim of this work was to study the role of the cell wall protein expansin in elongation growth. Expansins increase cell wall extensibility in vitro and are thought to be involved in cell elongation. Here, we studied the regulation of two tomato (Lycopersicon esculentum cv Moneymaker) expansin genes, LeExp2 and LeExp18, in rapidly expanding tissues. LeExp2 was strongly expressed in the elongation zone of hypocotyls and in the faster growing stem part during gravitropic stimulation. LeExp18 expression did not correlate with elongation growth. Exogenous application of hormones showed a substantial auxin-stimulation of LeExp2 mRNA in etiolated hypocotyls and a weaker auxin-stimulation of LeExp18 mRNA in stem tissue. Analysis of transcript accumulation revealed higher levels of LeExp2 and LeExp18 in light-treated, slow-growing tissue than in dark-treated, rapidly elongating tissue. Expansin protein levels and cell wall extension activities were similar in light- and dark-grown hypocotyl extracts. The results show a strong correlation between expansin gene expression and growth rate, but this correlation is not absolute. We conclude that elongation growth is likely to be controlled by expansin acting in concert with other factors that may limit growth under some physiological conditions.
Figures






References
-
- Ballas N, Wong L-M, Theologis A. Identification of the auxin-responsive element, AuxRE, in the primary indoleacetic acid-inducible gene, PS-IAA4/5, of pea (Pisum sativum) J Mol Biol. 1993;233:580–569. - PubMed
-
- Behringer FJ, Davies PJ. Indole-3-acetic acid levels after phytochrome-mediated changes in the stem elongation rate of dark- and light-grown Pisum seedlings. Planta. 1992;188:85–92. - PubMed
-
- Brummell DA, Bird CR, Schuch W, Bennett AB. An endo-1,4-β-glucanase expressed at high levels in rapidly expanding tissue. Plant Mol Biol. 1997;33:87–95. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

