Synthesis and posttranslational regulation of pyruvate formate-lyase in Lactococcus lactis
- PMID: 10940018
- PMCID: PMC111354
- DOI: 10.1128/JB.182.17.4783-4788.2000
Synthesis and posttranslational regulation of pyruvate formate-lyase in Lactococcus lactis
Abstract
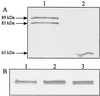
The enzyme pyruvate formate-lyase (PFL) from Lactococcus lactis was produced in Escherichia coli and purified to obtain anti-PFL antibodies that were shown to be specific for L. lactis PFL. It was demonstrated that activated L. lactis PFL was sensitive to oxygen, as in E. coli, resulting in the cleavage of the PFL polypeptide. The PFL protein level and its in vivo activity and regulation were shown by Western blotting, enzyme-linked immunosorbent assay, and metabolite measurement to be dependent on the growth conditions. The PFL level during anaerobic growth on the slowly fermentable sugar galactose was higher than that on glucose. This shows that variation in the PFL protein level may play an important role in the regulation of metabolic shift from homolactic to mixed-acid product formation, observed during growth on glucose and galactose, respectively. During anaerobic growth in defined medium, complete activation of PFL was observed. Strikingly, although no formate was produced during aerobic growth of L. lactis, PFL protein was indeed detected under these conditions, in which the enzyme is dispensable due to the irreversible inactivation of PFL by oxygen. In contrast, no oxygenolytic cleavage was detected during aerobic growth in complex medium. This observation may be the result of either an effective PFL deactivase activity or the lack of PFL activation. In E. coli, the PFL deactivase activity resides in the multifunctional alcohol dehydrogenase ADHE. It was shown that in L. lactis, ADHE does not participate in the protection of PFL against oxygen under the conditions analyzed. Our results provide evidence for major differences in the mechanisms of posttranslational regulation of PFL activity in E. coli and L. lactis.
Figures


Similar articles
-
The level of pyruvate-formate lyase controls the shift from homolactic to mixed-acid product formation in Lactococcus lactis.Appl Microbiol Biotechnol. 2002 Mar;58(3):338-44. doi: 10.1007/s00253-001-0892-5. Epub 2001 Dec 8. Appl Microbiol Biotechnol. 2002. PMID: 11935185
-
Cloning, expression, and characterization of the Lactococcus lactis pfl gene, encoding pyruvate formate-lyase.J Bacteriol. 1997 Sep;179(18):5884-91. doi: 10.1128/jb.179.18.5884-5891.1997. J Bacteriol. 1997. PMID: 9294449 Free PMC article.
-
Molecular characteristics and transcription of the gene encoding a multifunctional alcohol dehydrogenase in relation to the deactivation of pyruvate formate-lyase in the ruminal bacterium Streptococcus bovis.Arch Microbiol. 2004 Feb;181(2):122-8. doi: 10.1007/s00203-003-0638-0. Epub 2003 Dec 16. Arch Microbiol. 2004. PMID: 14676990
-
A radical-chemical route to acetyl-CoA: the anaerobically induced pyruvate formate-lyase system of Escherichia coli.FEMS Microbiol Rev. 1990 Aug;6(4):383-98. doi: 10.1111/j.1574-6968.1990.tb04108.x. FEMS Microbiol Rev. 1990. PMID: 2248795 Review.
-
A glycyl radical solution: oxygen-dependent interconversion of pyruvate formate-lyase.Mol Microbiol. 1998 Aug;29(4):945-54. doi: 10.1046/j.1365-2958.1998.00941.x. Mol Microbiol. 1998. PMID: 9767563 Review.
Cited by
-
A Novel Method for Long-Term Analysis of Lactic Acid and Ammonium Production in Non-growing Lactococcus lactis Reveals Pre-culture and Strain Dependence.Front Bioeng Biotechnol. 2020 Oct 8;8:580090. doi: 10.3389/fbioe.2020.580090. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 33163481 Free PMC article.
-
Glycolysis and the regulation of glucose transport in Lactococcus lactis spp. lactis in batch and fed-batch culture.Microb Cell Fact. 2007 May 24;6:16. doi: 10.1186/1475-2859-6-16. Microb Cell Fact. 2007. PMID: 17521452 Free PMC article.
-
Metabolic behavior of Lactococcus lactis MG1363 in microaerobic continuous cultivation at a low dilution rate.Appl Environ Microbiol. 2001 Jun;67(6):2677-82. doi: 10.1128/AEM.67.6.2677-2682.2001. Appl Environ Microbiol. 2001. PMID: 11375180 Free PMC article.
-
Metabolomic and proteomic insights into carbaryl catabolism by Burkholderia sp. C3 and degradation of ten N-methylcarbamates.Biodegradation. 2013 Nov;24(6):795-811. doi: 10.1007/s10532-013-9629-2. Epub 2013 Mar 5. Biodegradation. 2013. PMID: 23463356 Free PMC article.
-
13C based proteinogenic amino acid (PAA) and metabolic flux ratio analysis of Lactococcus lactis reveals changes in pentose phosphate (PP) pathway in response to agitation and temperature related stresses.PeerJ. 2017 Jul 5;5:e3451. doi: 10.7717/peerj.3451. eCollection 2017. PeerJ. 2017. PMID: 28695065 Free PMC article.
References
-
- Bolotin A, Mauger S, Malarme K, Ehrlich S D, Sorolin A. Low-redundancy sequencing of the entire Lactococcus lactis IL1403 genome. Antonie Leeuwenhoek. 1999;76:27–76. - PubMed
-
- Boumerdassi H, Desmazeaud M, Monnet C, Boquien C Y, Corrieu C. Improvement of diacetyl production by Lactococcus lactis ssp. lactis CNRZ 483 through oxygen control. J Dairy Sci. 1996;79:775–781.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

