Genetic evidence that transcription activation by RhaS involves specific amino acid contacts with sigma 70
- PMID: 10940041
- PMCID: PMC111377
- DOI: 10.1128/JB.182.17.4959-4969.2000
Genetic evidence that transcription activation by RhaS involves specific amino acid contacts with sigma 70
Abstract
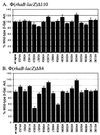
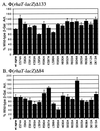
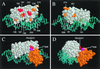
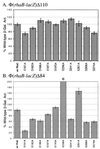
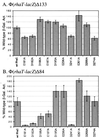
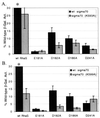
RhaS activates transcription of the Escherichia coli rhaBAD and rhaT operons in response to L-rhamnose and is a member of the AraC/XylS family of transcription activators. We wished to determine whether sigma(70) might be an activation target for RhaS. We found that sigma(70) K593 and R599 appear to be important for RhaS activation at both rhaBAD and rhaT, but only at truncated promoters lacking the binding site for the second activator, CRP. To determine whether these positively charged sigma(70) residues might contact RhaS, we constructed alanine substitutions at negatively charged residues in the C-terminal domain of RhaS. Substitutions at four RhaS residues, E181A, D182A, D186A, and D241A, were defective at both truncated promoters. Finally, we assayed combinations of the RhaS and sigma(70) substitutions and found that RhaS D241 and sigma(70) R599 met the criteria for interacting residues at both promoters. Molecular modeling suggests that sigma(70) R599 is located in very close proximity to RhaS D241; hence, this work provides the first evidence for a specific residue within an AraC/XylS family protein that may contact sigma(70). More than 50% of AraC/XylS family members have Asp or Glu at the position of RhaS D241, suggesting that this interaction with sigma(70) may be conserved.
Figures


References
-
- Akimuru H, Sakumi K, Yoshikai T, Anai M, Sekiguchi M. Positive and negative regulation of transcription by a cleavage product of Ada protein. J Mol Biol. 1990;216:261–273. - PubMed
-
- Baikalov I, Schroder I, Kaczor-Grzeskowiak M, Grzeskowaik K, Gunsalus R P, Dickerson R E. Structure of the Escherichia coli response regulator NarL. Biochemistry. 1996;35:11053–11061. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

