Analysis of topoisomerase function in bacterial replication fork movement: use of DNA microarrays
- PMID: 10944214
- PMCID: PMC16879
- DOI: 10.1073/pnas.97.17.9419
Analysis of topoisomerase function in bacterial replication fork movement: use of DNA microarrays
Abstract
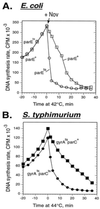
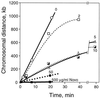
We used DNA microarrays of the Escherichia coli genome to trace the progression of chromosomal replication forks in synchronized cells. We found that both DNA gyrase and topoisomerase IV (topo IV) promote replication fork progression. When both enzymes were inhibited, the replication fork stopped rapidly. The elongation rate with topo IV alone was 1/3 of normal. Genetic data confirmed and extended these results. Inactivation of gyrase alone caused a slow stop of replication. Topo IV activity was sufficient to prevent accumulation of (+) supercoils in plasmid DNA in vivo, suggesting that topo IV can promote replication by removing (+) supercoils in front of the chromosomal fork.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

