Myosin-V stepping kinetics: a molecular model for processivity
- PMID: 10944217
- PMCID: PMC16890
- DOI: 10.1073/pnas.97.17.9482
Myosin-V stepping kinetics: a molecular model for processivity
Abstract
Myosin-V is a molecular motor that moves processively along its actin track. We have used a feedback-enhanced optical trap to examine the stepping kinetics of this movement. By analyzing the distribution of time periods separating discrete approximately 36-nm mechanical steps, we characterize the number and duration of rate-limiting biochemical transitions preceding each such step. These data show that myosin-V is a tightly coupled motor whose cycle time is limited by ADP release. On the basis of these results, we propose a model for myosin-V processivity.
Figures


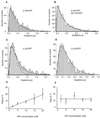
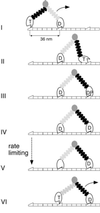
Comment in
-
A large step for myosin.Proc Natl Acad Sci U S A. 2000 Aug 15;97(17):9357-9. doi: 10.1073/pnas.97.17.9357. Proc Natl Acad Sci U S A. 2000. PMID: 10944206 Free PMC article. No abstract available.
References
-
- Cheney R E, O'Shea M K, Heuser J E, Coelho M V, Wolenski J S, Espreafico E M, Forscher P, Larson R E, Mooseker M S. Cell. 1993;75:13–23. - PubMed
-
- Reck-Peterson S L, Provance D W, Jr, Mooseker M S, Mercer J A. Biochim Biophys Acta. 2000;1496:36–51. - PubMed
-
- Howard J. Nature (London) 1997;389:561–567. - PubMed
-
- Mehta A D, Rock R S, Rief M, Spudich J A, Mooseker M S, Cheney R E. Nature (London) 1999;400:590–593. - PubMed
-
- Hackney D D. Annu Rev Physiol. 1996;58:731–750. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

